ABSTRACT
In response to nutrient starvation, the budding yeast Saccharomyces cerevisiae abandons mitotic proliferation and embarks on a differentiation process that leads through meiosis to the formation of haploid spores. This process is driven by cascading waves of meiosis-specific-gene expression. The early meiosis-specific genes are repressed during mitotic proliferation by the DNA-binding protein Ume6 in combination with repressors Rpd3 and Sin3. The expression of meiosis-specific transcription factor Ime1 leads to activation of the early meiosis-specific genes. We investigated the stability and promoter occupancy of Ume6 in sporulating cells and determined that it remains bound to early meiosis-specific gene promoters when those genes are activated. Furthermore, we find that the repressor Rpd3 remains associated with Ume6 after the transactivator Ime1 has joined the complex and that the Gcn5 and Tra1 components of the SAGA complex bind to the promoter of IME2 in an Ime1-dependent fashion to induce transcription of the early meiosis-specific genes. Our investigation supports a model whereby Ume6 provides a platform allowing recruitment of both activating and repressing factors to coordinate the expression of the early meiosis-specific genes in Saccharomyces cerevisiae.
INTRODUCTION
Modulation of gene expression is crucial to cellular development and differentiation processes where large sets of genes must be activated and deactivated in a tightly orchestrated fashion to achieve changes in cell phenotype and morphology. Coordinated assembly, modification, and disassembly of transcription factor complexes are major mechanisms that can be employed to control regulated transcription (Citation1). Gamete formation (sporulation) in Saccharomyces cerevisiae is a well-studied developmental process that is driven by temporally regulated waves of gene expression (Citation2). Upon nitrogen and glucose starvation, S. cerevisiae abandons mitotic proliferation and initiates a differentiation program that leads through meiosis to the formation of haploid spores. The combination of nutrient signaling and MATa/MATα genome status triggers a cascade of gene expression that activates previously silent genes and promotes progression through premeiotic DNA replication, elevated rates of homologous recombination, reductional and equational chromosome division, and finally, spore morphogenesis (Citation3Citation–Citation5). An immediate response to starvation is induction of the Ime1 transcription factor, which has been referred to as the “master regulator” of meiosis and the sporulation process (Citation6). IME1 is strictly required for the induction of early meiosis-specific-gene expression (Citation7Citation–Citation9).
The early gene family consists of genes whose products function to promote and regulate early events of the sporulation program, including premeiotic DNA replication, homologous recombination, and progression into the meiotic chromosome divisions (Citation3, Citation5). Most of these genes are held silent during mitotic proliferation. A major component of the regulation of these genes is imposed through the conserved URS1 DNA sequence found upstream from the open reading frames of all members of this gene family (Citation10). The URS1 DNA sequence is a binding site for the Ume6 protein (Citation11). During mitotic proliferation, Ume6 bound to URS1 recruits histone deacetylase Rpd3, corepressor Sin3, and chromatin remodeling factor Isw2 (Citation12, Citation13). These factors repress early meiosis-specific-gene expression in part through deacetylation of the surrounding histones and formation of inaccessible chromatin structures (Citation14Citation–Citation16). Mutational inactivation of Ume6, Rpd3, or Sin3 results in derepression of early meiosis-specific genes, allowing their expression during vegetative growth (Citation17Citation–Citation19). These observations led to the proposal that Ume6 acts as a repressive factor to control early meiosis-specific genes. Induction of the early meiosis-specific genes is dependent upon Ime1 expression and nuclear localization (Citation7, Citation20). Ime1 has a transcriptional activation domain and transcriptional activation capabilities when tethered to DNA but displays no intrinsic sequence-specific DNA binding capability (Citation9, Citation21). Ime1 is not associated with early meiosis-specific gene promoters in ume6 mutant strains, suggesting that Ume6 recruits the Ime1 trans activator to trigger early gene expression during sporulation (Citation22, Citation23). Two-hybrid experiments revealed that Ime1 can bind Ume6 and that mutations in threonine 99 of Ume6 block this interaction (Citation22). Additionally, Ime1 mutants that can bind to Ume6 but lack the sequences encoding the transcription-activating domain are defective in inducing meiosis-specific-gene expression (Citation21). The observation that a LexA-Ume6 fusion protein confers Ime1-dependent activation on a LexA operator-regulated reporter gene is consistent with the proposal that Ume6 represses meiosis-specific genes through recruitment of Sin3-Rpd3 repressors and that the complex is converted into an activator upon binding Ime1 (Citation22, Citation24). This parsimonious model is further supported by the observation that a Gal4 activation domain (GAD)-Ume6 fusion protein allows meiosis in cells lacking Ime1 (Citation24).
A competing model for regulation of early meiosis-specific genes was proposed by Mallory et al., who showed that Ume6 was degraded in an Ime1-dependent fashion, removing the repressive complex from early gene promoters (Citation25). It was further proposed that Ume6 destruction and early meiosis-specific-gene expression was dependent upon the Cdc20-activating subunit of the anaphase promoting complex (APCCdc20) (Citation25). This model provided no mechanism for meiosis-specific induction of genes beyond the derepressed level achieved by loss of Ume6.
Cdc20 performs essential functions during mitotic proliferation and during progress through meiosis (Citation26). During the sporulation process, Cdc20 is expressed in the middle phase of meiosis, and cells subjected to meiosis-specific depletion of Cdc20 arrest at metaphase I, displaying metaphase I spindles, complete DNA replication, and high levels of Pds1 (Citation26, Citation27). Proteins encoded by early genes REC8 and HOP1 and middle genes CLB5, NDT80, and CLB1 accumulate with normal kinetics in sporulating cells depleted of Cdc20 (Citation28). Thus, it is surprising that Cdc20 should be implicated in triggering the expression of the early meiosis-specific genes. The APC subunit Cdh1 is expressed during sporulation but is not critical for progression through sporulation and spore formation (Citation27, Citation29, Citation30). In contrast, Ama1 is a meiosis-specific APC activating subunit (Citation31). Cells lacking Ama1 progress into the early phase of the sporulation program with wild-type kinetics (Citation27, Citation32, Citation33), although some strain backgrounds display defects in chromosome divisions and spore wall defects (Citation34, Citation35). In contrast to the lack of a demonstrable requirement for APC activity to allow progression into and through the early phase of sporulation, protein degradation by autophagy is essential for sporulation (Citation36, Citation37). Cells deficient in autophagy factor Atg12 arrest prior to meiotic DNA replication and lose viability (Citation37). Ume6 is not a substrate for degradation by the autophagy process in S. cerevisiae. Indeed, Ume6 is a regulator of the gene encoding the essential autophagy factor Atg8 (Citation36).
The lysine acetyltransferase Gcn5 is a component of the SAGA and SLIK complexes, which interact with transcriptional activators to promote active transcription (Citation38, Citation39). Gcn5 is also required for the induction of meiosis-specific genes and for sporulation (Citation40). However, previous studies have not demonstrated that Gcn5 or SAGA specifically associates with meiosis-specific transcriptional activators. Histones surrounding the URS1 sites in the IME2 promoter display a Gcn5-dependent increase in acetylation, suggesting that SAGA may be recruited to this promoter, but it has also been proposed that Gcn5 acetylates Ume6 to promote Ume6’s destruction (Citation41).
Activation of the early meiosis-specific genes by destruction of Ume6 is inconsistent with a requirement for Ume6 to recruit Ime1 in the role of a trans activator. Additionally, the failure of ume6 deletion strains to effectively induce early meiosis-specific genes and progress through the sporulation program is inconsistent with a purely repressive role for Ume6 (Citation42). Our investigation of early meiosis-specific-gene regulation suggests that Ume6 remains bound to the promoters of early meiosis-specific genes IME2 and SPO13 throughout the early stages of the sporulation program, where it creates a platform for the recruitment of both transcriptional repressor Rpd3 and activator Ime1.
RESULTS
Ume6 is stable during the early phase of the sporulation program.
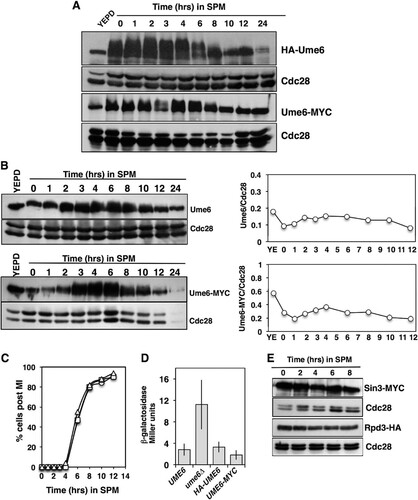
We monitored the abundance of hemagglutinin-tagged Ume6 (HA-Ume6) in a population of cells induced to synchronously initiate sporulation. Protein extracts prepared under denaturing conditions and analyzed by Western blotting revealed that the HA-Ume6 abundance did not display a significant reduction during the early phase of sporulation from 0 to 8 h (). Ume6 did display slow mobility species as the cells initiated the sporulation program (0 to 6 h). This reduced mobility has been observed by others and was largely attributed to phosphorylation of Ume6 by kinases Rim11 and Rim15 (Citation43). Some reduction in the Ume6 abundance was noted in the later phases of sporulation; however, during the time period in which early genes are activated (1 to 4 h), there did not appear to be a dramatic reduction in the Ume6 abundance. The HA-tagged version of Ume6 was functional in that HA-UME6 cells displayed sporulation timing and frequency similar to those of UME6 cells (). Additionally, HA-UME6 was able to repress an IME2-LacZ reporter gene in actively proliferating cells to an extent similar to that observed with UME6 (). A second version of Ume6, with a carboxyl-terminal MYC tag, was also generated. In diploid cells induced to initiate the sporulation program, Ume6-MYC displayed behavior similar to that of HA-Ume6 (). The abundance of Ume6-MYC was not reduced during the early phase of sporulation, but Ume6-MYC did display more slowly migrating species that were not apparent during the later phases of sporulation (). Like HA-Ume6, Ume6-MYC was functional, based upon the ability of UME6-MYC-expressing cells to undergo meiosis and sporulation with timing and frequency similar to those of a UME6 strain and to enforce repression of an IME2-LacZ reporter gene during mitotic proliferation ( and ). Although the added HA or MYC epitope tags did not appear to alter Ume6 function with regard to progression through the sporulation program, we considered that the appended epitope tag sequences may have influenced Ume6’s stability during this process. To test this possibility, protein extracts were prepared from a strain expressing the native untagged Ume6 (DSY1089) and subjected to Western blot analysis using an anti-Ume6 antibody directed against residues 339 to 808. This analysis revealed that the native Ume6 protein displayed a pattern of abundance similar to those displayed by the epitope-tagged versions of Ume6 (). A Li-COR Odyssey scanner was used to quantify the Western blot signals for this experiment, and the Ume6/Cdc28 ratios were plotted (). This analysis revealed that the Ume6 abundance was higher in cells proliferating in yeast extract-peptone-dextrose (YEPD) medium and that there was some fluctuation in Ume6 abundance throughout the sporulation program but no dramatic reduction during the early and middle periods for sporulation (2 to 8 h), when early meiosis-specific genes are induced. A similar result was observed when monitoring Ume6-MYC (, bottom).
FIG 1 Ume6 protein abundance does not decrease during the early phase of sporulation. (A) Western blots detecting the abundance of HA-Ume6 (top) and Ume6-MYC (bottom) during vegetative growth (YEPD) or following induction of the sporulation program in sporulation medium (SPM). Cdc28 abundance was monitored as a control for protein loading. Ume6 and Cdc28 were probed on the same membranes. (B) The signals from untagged Ume6 probed with an anti-Ume6 antibody, Ume6-MYC probed with anti-MYC antibody, and loading control Cdc28 were quantified and are presented as the Ume6/Cdc28 ratios. (C) Percentages of diploid UME6/UME6 (circles), HA-UME6/HA-UME6 (squares), and UME6-MYC/UME6-MYC (triangles) cells displaying two or more segregated chromatin masses following inoculation into SPM. (D) β-Galactosidase activity produced in actively proliferating cells harboring a centromeric IME2-LacZ reporter plasmid. Activity is reported in Miller units. Data are reported as the mean levels of activity measured from three independent transformants. The error bars reflect standard deviations. (E) Western blot analysis of Sin3-MYC, Rpd3-HA, and Cdc28 abundance in cultures induced to initiate synchronous sporulation by inoculation into SPM. Sin3-MYC samples were separated on a 6% SDS–polyacrylamide gel, and duplicate samples were separated on a 10% gel to probe for Cdc28. Rpd3-HA samples were electrophoresed on duplicate 10% SDS–polyacrylamide gels; one was probed for Rpd3-HA and a duplicate probed for Cdc28.
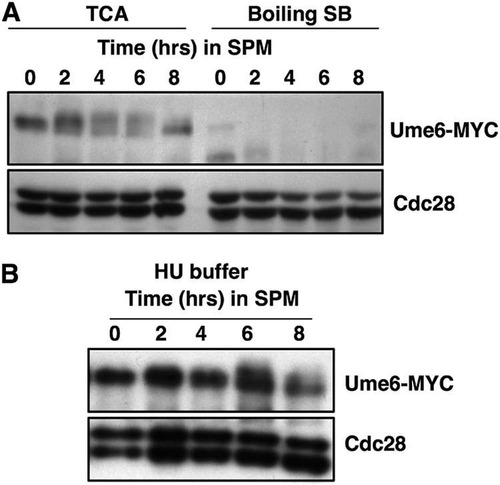
As these findings differ from those reported previously, we investigated the abundance of Ume6 that could be detected when the protein extracts were prepared by boiling cell pellets in SDS sample buffer (Citation25, Citation44). When protein extracts were prepared by vortexing with glass beads in trichloroacetic acid (TCA) followed by precipitation, as described in Materials and Methods, Ume6-MYC could be detected during the 2 to 8 h following induction of the sporulation program (, TCA). In contrast, when protein samples were prepared by boiling cell pellets in SDS sample buffer, the signal for Ume6-MYC was specifically reduced (, Boiling SB). A similar reduction in the Ume6 signal was observed when the protein extracts were prepared by mechanical lysis under nondenaturing conditions (data not shown). Knop et al. have described a method to lyse cells in NaOH, followed by TCA precipitation and solubilization in buffers containing urea and SDS (Citation45). This method has been reported to efficiently extract proteins from sporulating cells. A much lower reduction in the Ume6-MYC signal was observed when extracts were prepared with this method than from boiling cells in SDS sample buffer ().
FIG 2 Denaturing extraction with TCA improves the detection of Ume6-MYC over extraction by boiling in SDS sample buffer. Samples of UME6-MYC/UME6-MYC cells were collected at the indicated time points following the induction of sporulation. (A) Protein extracts were prepared by either vortexing cell pellets with glass beads in 20% trichloroacetic acid (TCA) or by boiling in 1× SDS sample buffer (SB). (B) Cells harvested at the indicated time points following induction of sporulation were lysed with NaOH and the proteins precipitated by addition of TCA prior to resuspending in 8 M urea–5% SDS. Samples were probed by Western blotting for Ume6-MYC and Cdc28 as indicated.
These observations suggest that, in vivo, Ume6 is not dramatically degraded during the early phase of the sporulation program but that it may be labile upon cell lysis or difficult to extract from sporulating cells. The relative stability of Ume6 that we observed is consistent with earlier findings that Ume6 not only performs an important role for imposing repression on early meiosis-specific genes during vegetative growth but is also required for the effective induction of this gene family during sporulation.
Repressors Sin3 and Rpd3 are stable throughout the sporulation process.
Ume6 imposes repression on meiosis-specific genes by forming a complex with histone deacetylase Rpd3 and corepressor Sin3. To determine whether or not the two repressive factors were degraded upon entry into and progression through sporulation, we monitored the abundance of Sin3-MYC and Rpd3-HA during the course of a synchronous sporulation (). Sin3-MYC was present in actively proliferating cells (T = 0) and could also be detected during the early portion of the sporulation time course, up to 8 h, when the early meiosis-specific genes are induced. During the early phase of sporulation, the abundance of Sin3-MYC remained relatively constant (, time points 0 to 8 h). Similarly, Rpd3-HA could be detected as the cells progressed through the sporulation process (). There was little reduction in Sin3-MYC or Rpd3-HA through the 2- to 6-h time points, when the early meiosis-specific genes are induced ().
Cdc20-associated APC activity is not required for induction of the early meiosis-specific genes.
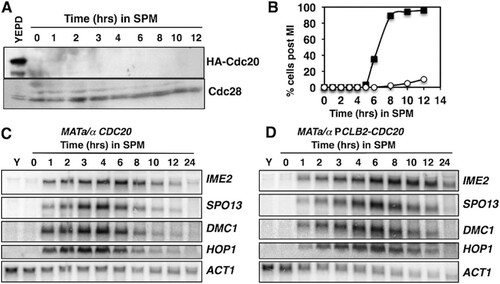
It has been proposed that APCCdc20 regulates early meiosis-specific-gene expression through control of Ume6’s stability (Citation25). Although our data suggest that the abundance of Ume6 does not decrease significantly in the early phases of sporulation, we investigated whether APCCdc20 had a role in regulating early meiosis-specific-gene expression. CDC20 is an essential gene, so to test its effects on Ume6’s stability and early meiosis-specific-gene expression, we made use of a CDC20 allele in which the open reading frame is placed under the regulation of the CLB2 promoter, which is expressed exclusively during mitotic proliferation (Citation26). These cells effectively expressed Cdc20 during mitotic proliferation (, YEPD), but upon completion of the mitotic M phase and entry into the sporulation program, Cdc20’s abundance was reduced and undetectable when cells were transferred into sporulation medium (, 0 to 12 h) (Citation26). This strain is reported to display premeiotic DNA replication with normal timing and formation of a metaphase spindle (Citation27). However, the strain undergoes arrest in meiosis I (MI) () (Citation26). Although the PCLB2-CDC20 strain displayed arrest in MI, we could observe no significant difference in the timing of induction or the abundance of transcripts of early genes IME2, SPO13, DMC1, and HOP1 ( and ). The abundance of the early gene transcripts declined as CDC20/CDC20 cells progressed through meiosis I (MI) and meiosis II (MII) (). In contrast, the early gene transcripts persisted for a longer time in the PCLB2-CDC20/PCLB2-CDC20 strain (), consistent with arrest of the strain at metaphase I (Citation27). These observations are inconsistent with a scenario where Cdc20-dependent destruction of Ume6 is required to release these genes from repression.
FIG 3 Cdc20 is not required for the induction of early meiosis-specific genes. (A) Western blot analysis of HA-Cdc20 in actively proliferating PCLB2-CDC20/PCLB2-CDC20 cells (YEPD) and in cells induced to initiate sporulation by inoculation into SPM. The upper half of the membrane was probed with an anti-HA antibody, and the lower portion of the membrane was probed for Cdc28 as a loading control. (B) Percentages of diploid CDC20/CDC20 (squares) and PCLB2-CDC20/PCLB2-CDC20 (circles) cells displaying two or more segregated DAPI-stained chromatin masses. (C, D) Northern blot analysis of transcripts of early meiosis-specific genes IME2, SPO13, DMC1, and HOP1. The ACT1 transcript was included as a control. RNA was collected from actively proliferating cultures (Y) or at the indicated time points following induction of sporulation.
Ume6 remains bound to early gene promoters throughout the early phase of sporulation.
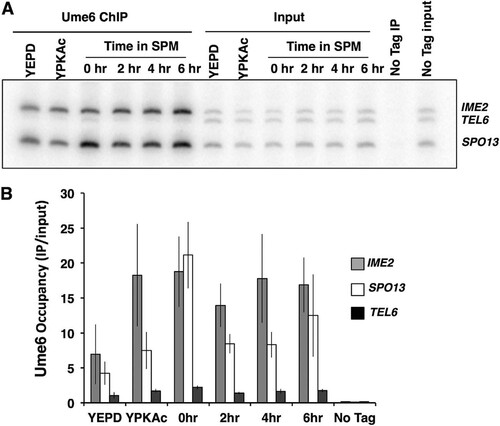
Although we observed that the Ume6 abundance was not reduced during the early phase of the sporulation program, we considered the possibility that DNA-bound Ume6 is specifically targeted or that Ume6 is displaced from early gene promoters. We investigated the occupancy of Ume6 at the promoters of the early meiosis-specific genes SPO13 and IME2, as well as the telomere of chromosome VI, which has no identifiable URS1 binding site for Ume6 (Citation46). Chromatin immunoprecipitation (ChIP) was performed to investigate the abundance of Ume6 present on these early meiosis-specific gene promoters. PCR analysis of Ume6-MYC immunoprecipitates from mitotically proliferating cells (YEPD medium), as well as cells in the early phase of sporulation (0 to 6 h), displayed enrichment for products amplified from the SPO13 and IME2 promoters relative to the TELVI signal, indicating occupancy of Ume6 on the early gene promoters. The SPO13 and IME2 product signals did not display any significant reduction during the early phase of the sporulation program, when SPO13 and IME2 mRNA accumulated maximally (). These observations are consistent with the proposal that Ume6 remains associated with early meiosis-specific gene promoters both when these genes are repressed and once they become activated.
FIG 4 Ume6 remains bound to early meiosis-specific gene promoters during sporulation. (A) PCR analysis of Ume6-MYC immunoprecipitates (Ume6 ChIP) or samples of the input extracts (Input) collected from cultures growing in medium supplemented with glucose (YEPD) or acetate (YPKAc) or from cells at the indicated times following inoculation into sporulation medium (SPM). The “No Tag IP” label is used for immunoprecipitate or input DNA from an untagged UME6 strain. (B) Quantitative analysis of the PCR products from Ume6-MYC immunoprecipitates. Data presented are mean IP/input ratios from three repetitions; error bars reflect standard deviations.
Chromatin-bound Ume6 binds both activators and repressors of meiosis-specific transcription.
Expression of the Ime1 transcription factor is required for induction of the early meiosis-specific genes (Citation7, Citation21). Ime1 displays no specific DNA binding activity of its own but can physically interact with Ume6 in a yeast two-hybrid assay, leading to the suggestion that Ume6 recruits Ime1 to early meiosis-specific promoters, thus allowing their activation (Citation7, Citation9, Citation22, Citation24). We investigated the association of Ime1 with Ume6 at early gene promoters using ChIP. Ume6-MYC protein-DNA complexes were immunoprecipitated and then reprecipitated using an anti-HA antibody recognizing Ime1-HA. Cells actively proliferating in rich growth medium (YEPD medium) displayed a very low signal for Ime1-HA at either of the early gene promoters IME2 and SPO13, consistent with the lack of expression of Ime1 in growing cells (). In contrast, cells growing in acetate medium (YEP medium supplemented with 1% potassium acetate [YPKAc]) displayed an Ime1-HA signal at the IME2 and SPO13 promoters with no detectable signal for the control TEL6 sequence. The occupancy of Ime1-HA at the IME2 and SPO13 promoters increased following induction of the sporulation program, reaching an approximately 50-times increase at the IME2 URS1 sequence after 3.5 h, at the time that early gene expression is induced ( and ). These data indicate that Ime1 and Ume6 are both resident on early meiosis-specific gene promoters at the time these genes are induced.
FIG 5 Ime1 and Rpd3 are bound to Ume6 during sporulation. (A) PCR analysis of Ume6-MYC immunoprecipitates that were reprecipitated with anti-HA antibodies to isolate Ime1-HA. Immunoprecipitate from a UME6-MYC IME1 strain (No tag) or UME6-MYC IME1-HA strain actively proliferating in medium supplemented with glucose (YEPD) or acetate (YPKAc) or induced to sporulate (SPM). (B) Quantitative analysis of the PCR products from Ume6-MYC Ime1-HA immunoprecipitates. (C) PCR analysis of Ume6-MYC immunoprecipitates that were reprecipitated with anti-HA antibodies to isolate Rpd3-HA. Immunoprecipitate from a UME6-MYC RPD3 strain (No tag) or UME6-MYC RPD3-HA strain actively proliferating in medium supplemented with glucose (YEPD) or acetate (YPKAc) or induced to sporulate (SPM). (D) Quantitative analysis of the PCR products from Ume6-MYC Rpd3-HA immunoprecipitates. Data presented are mean IP/input ratios from three repetitions; error bars reflect standard deviations.
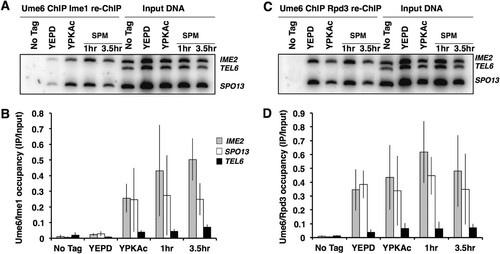
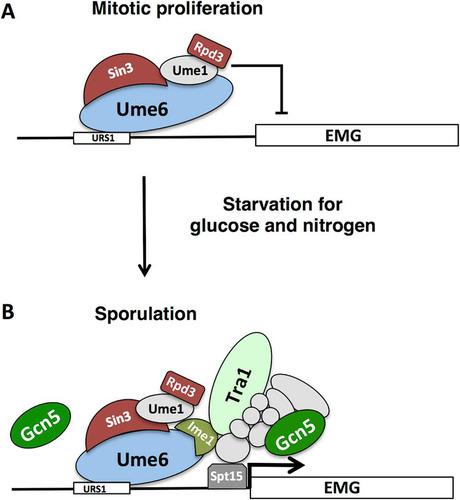
During vegetative growth of S. cerevisiae, the histone deacetylase Rpd3 and the corepressor Sin3, which are tethered to Ume6, impose repression on the early meiosis-specific gene family. Upon entry into the sporulation program, the early genes are activated, and we anticipated that the arrival of Ime1 might trigger the displacement of Rpd3 to allow activation of the meiosis-specific genes. As anticipated, sequential ChIP (ChIP re-ChIP) experiments with anti-MYC antibodies recognizing Ume6-MYC followed by anti-HA antibodies recognizing Rpd3-HA revealed that Rpd3-Ume6 complexes are present at the IME2 and SPO13 promoters in cultures proliferating in YEPD or YPKAc medium (). Surprisingly, the IME2 and SPO13 PCR signals from the Rpd3-Ume6 complexes were not reduced as the cultures were induced to initiate the sporulation program. Rpd3-Ume6 complexes remained associated with the IME2 and SPO13 promoters after 1 and 3.5 h following induction of sporulation, when IME2, SPO13, and other early meiosis-specific genes are strongly induced ( and ). ChIP performed with anti-Rpd3 antibodies has yielded similar results (Citation47). One explanation for this observation is that upon induction of the sporulation program, Ime1 binds to Ume6-Rpd3 complexes and is able to activate the target genes. Alternatively, it could be that in a portion of the cells, Ume6 retains Rpd3 and holds the early meiosis-specific genes in a repressed state, while Ime1 binds Ume6 to activate IME2, SPO13, and the other early genes in a distinct subset of the population. The latter explanation seems unlikely in this strain background, based on the high frequency and synchrony with which cells complete the sporulation process. These observations suggest not only that Ime1 binds to Ume6 coincident with the activation of early meiosis-specific genes but that complexes containing Ume6 and both Rpd3 and Ime1 can form on the early gene promoters.
Ime1 is required for Gcn5 occupancy of an early meiosis-specific promoter.
Gcn5 is required for induction of early meiosis-specific genes, and cells lacking functional Gcn5 display a defect in progression into the sporulation program (Citation40). Early meiosis-specific-gene induction is also dependent upon Ime1. To test the hypothesis that Gcn5 might be required for Ime1 to activate the reporter gene, LexA-Ime1 was expressed in either a GCN5 or a gcn5::natMX4 strain harboring a LexA operator-regulated LacZ reporter gene, and the levels of β-galactosidase activity were compared. The LexA-Ime1 fusion effectively induced the expression of the LacZ reporter gene, whereas LexA alone induced very little β-galactosidase activity (). Deletion of GCN5 reduced the ability of LexA-Ime1 to activate the reporter, as shown by the greater than 5-fold reduction in β-galactosidase activity detected (741 ± 165.6 [mean ± standard deviation] versus 137 ± 9.2; P = 0.0029) (). Deletion of GCN5 significantly reduced the expression of the LacZ reporter gene, suggesting that Ime1 may be required for Gcn5 to act at early meiosis-specific gene promoters to induce their activation.
TABLE 1 β-Galactosidase activity driven by LexA-Ime1 fusionTable Footnotea
To further investigate the possibility that Ime1 was required for Gcn5 to participate in the activation of early meiosis-specific gene promoters, we performed ChIP with MYC-tagged Gcn5 to determine whether Gcn5 was resident at the IME2 promoter at early times during the sporulation program when IME2 was actively transcribed. Quantitative PCR (qPCR) analysis of Gcn5-MYC immunoprecipitates revealed little enrichment for IME2 promoter DNA in cells mitotically proliferating in YEPD rich growth medium when IME2 is not expressed (). In contrast, IME2 promoter DNA was enriched more than 6-fold above its level in the no-antibody control in Gcn5-MYC immunoprecipitates prepared 3 h following the induction of the sporulation program (). No enrichment of IME2 promoter DNA could be detected when ChIP was performed with an untagged GCN5 strain, either during mitotic proliferation or sporulation (). If Ime1 were required for Gcn5 occupancy of IME2 promoter DNA during sporulation, we predicted that deletion of IME1 or UME6 would reduce the presence of Gcn5 at the IME2 promoter. This hypothesis was tested by performing ChIP analysis with GCN5-MYC/GCN5-MYC ime1/ime1 diploids that had been induced to initiate the sporulation program. While IME2 promoter DNA was readily detected in anti-MYC antibody immunoprecipitates from a GCN5-MYC/GCN5-MYC IME1/IME1 diploid (), the enrichment in the ime1/ime1 diploid was reduced to 2-fold above the level in the no-antibody control (). No enrichment of IME2 promoter DNA could be detected when the experiment was performed with strains expressing untagged GCN5. Similarly, ChIP analysis demonstrated reduced Gcn5 occupancy of the IME2 promoter in a ume6/ume6 strain 3 h after induction of the sporulation program (). The reduction in binding of Gcn5-MYC to the IME2 promoter appears to be greater in the ume6/ume6 strain than in the ime1/ime1 strain, implying that Ume6 or some component of the chromatin-bound Ume6 complex may promote binding of Gcn5 to the IME2 promoter in the absence of Ime1. However, the degree of variance among replicate experiments makes it difficult to quantitatively support that conclusion. The apparent reduced presence of Gcn5 at the IME2 promoter in mutants lacking either IME1 or UME6 is consistent with DNA-bound Ume6 acting as a platform that is required for both Ime1 and Gcn5 to occupy the IME2 promoter.
FIG 6 Gcn5 and Tra1 are present at the IME2 promoter during sporulation. Strains expressing untagged GCN5 or GCN5-MYC (A to C) or untagged TRA1 or HA-TRA1 (D to E) during vegetative growth (YEPD) or 3 h post-induction of sporulation (SPM) were subjected to ChIP using anti-MYC or anti-HA antibodies. The fold increase reflects the increase in the IME2 URS1 PCR product relative to the negative-control TEL6 sequence that does not harbor a URS1 sequence. (A, D) Wild type. (B, E) ime1/ime1. (C) ume6/ume6. The data presented reflect mean values from three independent ChIP replicates and three technical replicates of each sample; errors bars indicate standard deviations.
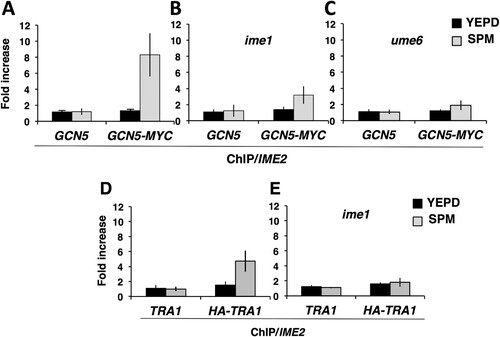
Tra1 binds to early meiosis-specific gene promoters and may link SAGA to the Ime1 transcriptional activator.
Gcn5 is a component of the 19-subunit SAGA complex that promotes gene activation (Citation48). The Tra1 subunit of this complex directly binds to transcriptional activators Gal4 and Gcn4 and thus brings Gcn5 and other SAGA subunits to the cognate promoters (Citation49, Citation50). We reasoned that Gcn5 might occupy the IME2 promoter as a component of the SAGA complex and tested the possibility that Tra1 bound to the Ume6-Ime1 complex on the IME2 promoter. ChIP analysis was performed with an HA-TRA1/HA-TRA1 strain. Anti-HA antibody immunoprecipitates from an untagged TRA1/TRA1 strain revealed no enrichment for the IME2 promoter DNA in either mitotically proliferating cells or sporulating cells (). In contrast, anti-HA immunoprecipitates from an HA-TRA1/HA-TRA1 strain displayed a significant increase in the presence of IME2 promoter DNA in extracts made from sporulating cells but not in extracts from mitotically proliferating cells (). To further test the hypothesis that DNA-bound Ume6 binds Ime1 and that Ime1 is required for Tra1 binding to the IME2 promoter, ChIP was performed on extracts from an HA-TRA1/HA-TRA1 ime1/ime1 strain or an untagged TRA1/TRA1 ime1/ime1 strain 3 h after induction of the sporulation program. No enrichment for IME2 promoter DNA was detected in immunoprecipitates from extracts of proliferating cultures ( and ). Similarly, there was no enrichment of IME2 promoter DNA associated with the HA-TRA1 immunoprecipitated from an ime1/ime1 strain compared to the amount from the no-antibody control (). Collectively these data support a model whereby DNA-bound Ume6 binds to meiosis-specific activator Ime1, and this in turn is required for Tra1 and SAGA to occupy the IME2 promoter and, possibly, the promoters of other early meiosis-specific genes ().
FIG 7 Regulation of early meiosis-specific genes. (A) During mitotic proliferation, early meiosis-specific genes (EMG) are repressed through the action of Ume6 binding to the URS1 DNA sequence. Ume6 acts as a platform to tether repressors Sin3, Rpd3 and other accessory proteins to targeted promoters. (B) Upon starvation for glucose and nitrogen, Ime1 is expressed and binds to Ume6. Our data suggest that Ume6 remains bound to the URS1 sequences of early meiosis-specific genes IME2 and SPO13 and that Rpd3 remains associated with Ume6 following Ime1 binding. Interaction of Ime1 with Ume6 is essential for the induction of EMGs in a Gcn5-dependent fashion. Tra1 binds the IME2 promoter in an Ime1-dependent fashion, raising the possibility that Ime1 directly binds to Tra1, possibly within the context of the SAGA complex. Gcn5 is essential for the induction of meiosis-specific genes and may function in conjunction with Tra1 as a part of the SAGA complex that aids in loading the TATA box binding protein Spt15. Gcn5 may also act directly on Ume6, but it is unclear whether it does so as a part of SAGA or through a distinct protein complex.
DISCUSSION
Ume6 remains associated with early meiosis-specific gene promoters during the early phase of sporulation.
The data presented here support a model whereby Ume6 provides a DNA binding platform that directs both positively and negatively acting factors to regulate the expression of early meiosis-specific genes (Citation22, Citation24). Binding of Sin3 and Rpd3 provides a repressive complex that regulates the chromatin modification state and, possibly, the acetylation state of Ume6 and other proteins in the complex (Citation14, Citation41). Starvation for nitrogen and glucose induces the expression of Ime1. Modification of Ime1 and Ume6 by Rim11 kinase then promotes association between Ume6 and Ime1, allowing Ime1 to activate the meiosis-specific target genes. Our data support a model whereby this activation is accomplished at least in part through recruitment of Tra1 and the SAGA complex. Based on Western blot analysis from samples prepared under denaturing conditions and on ChIP data, we conclude that Ume6 remains associated with early gene promoters through the early phases of sporulation when early meiosis-specific genes are induced. The repressive function of Ume6 is well documented, and deletion of Ume6 allows derepression of meiosis-specific genes during vegetative growth (Citation17). In contrast, cells lacking Ume6 cannot further induce the expression of early meiosis-specific genes upon starvation and cannot progress effectively into the sporulation program, supporting the contention that Ume6 is required for effective induction of the genes it regulates. Consistent with this is the observation that mutation of URS1, the Ume6 binding site, prevents induction of meiosis-specific genes during sporulation (Citation17, Citation22). Furthermore, Ime1 is strictly required for induction of the early gene family, and Ime1 binds to Ume6 (Citation17). Indeed, Ime1’s recruitment to early gene promoters and induction of those genes is dependent upon Ume6 (Citation23). The addition of a Gal4 transcriptional activation domain to Ume6 (GAD-Ume6) does not overcome repression of early meiosis-specific genes in vegetative cells, but in cells lacking the repressor Sin3 or Rpd3, GAD-Ume6 can activate a URS1-regulated reporter gene (Citation51). The GAD-Ume6 fusion can rescue the sporulation defect in Ime1-deficient diploids, although not to 100% (Citation24, Citation51). Further supporting a positive role for Ume6 in meiosis-specific-gene activation is the observation that LexA-Ume6 fusions can activate LexA operator-regulated reporter genes, but only if Ime1 is expressed (Citation22). These observations, in addition to the data reported here, are not consistent with a model where destruction of Ume6 is required to allow activation of the regulated genes. Rather they support a model where Ume6 acts as a binding platform for both repressive factors and the activator Ime1.
It is not entirely clear why our findings differ from previously published observations that Ume6 is rapidly degraded during the early phase of sporulation. In agreement with previous investigations, we observe that Ume6 is more abundant in cells proliferating in rich glucose medium than in cells in acetate medium, but Ume6 is present throughout the early phase of sporulation. Variables that may have yielded some of these differences include the use of different S. cerevisiae strain backgrounds. We made use of SK1 strains, whereas Mallory et al. performed their investigation with an SK1/W303 hybrid (Citation25). Additionally, we made use of different epitope tags and performed protein extractions using a different method. We observe that in vegetative cells, Ume6 is readily detectable by any of the extraction methods performed, but upon initiating sporulation, Ume6 is more difficult to reliably detect with native extraction procedures or by simply boiling cell pellets in SDS sample buffer. It is unclear whether this reflects decreased stability or solubility in the extracts. It may be relevant that in the original description of the method for protein extraction by boiling in SDS sample buffer, the authors noted that protein extraction was much less efficient from cells grown in minimal medium than from cultures grown in rich medium (Citation52).
APCCdc20 activity is not required for induction of early meiosis-specific genes.
We find no evidence to support a role for APC subunit Cdc20 in the degradation of Ume6 during the early phase of sporulation or induction of the early meiosis-specific genes. Cdc20 accumulates in the middle phase of sporulation rather than at early times, and it serves an essential role in the completion of metaphase I (Citation26, Citation27). The role of Cdc20 at metaphase I is likely dependent on the proteosome, since inhibition with MG132 yields a similar metaphase I arrest without impeding progression through the early phases of sporulation (Citation37). We observed that meiosis-specific depletion of Cdc20 yielded the expected metaphase I arrest but did not alter the induction of the early meiosis-specific genes. While it is possible that sufficient Cdc20 remained in the cells to execute some function related to gene expression, we did not observe any effects on Ume6.
In contrast, autophagy is required for the early stages of sporulation, and Ume6 is a regulator of ATG8, an essential component of the autophagy system (Citation36). While there is no evidence that autophagy leads to destruction of Ume6, the precise role of autophagy in S. cerevisiae sporulation has not been determined. It may be that the process is required to provide precursors for nascent protein synthesis, since the sporulation process occurs in the absence of nitrogen and amino acids.
Although we did not see a dramatic reduction in the Ume6 abundance in the early phase of sporulation (0 to 4 h), we did observe reduction of Ume6 at late times in sporulation where Cdc20 may be more active; however, we have not investigated whether this is dependent on Cdc20 or the putative destruction box sequences in Ume6. This study employed a Cdc20 depletion technique, whereas a previous investigation made use of a thermosensitive cdc20-1 allele, which increases the complexity of the experiment since some aspects of sporulation are temperature sensitive in most S. cerevisiae strains (Citation25, Citation53). The difference in methods applied to inactivate Cdc20 may be responsible for some of the differences from our results.
It is entirely possible that Ume6 is targeted for degradation under some conditions or at some gene promoters since it regulates genes that are required for functions other than sporulation (Citation54). Thus, destruction of Ume6 within some contexts may be important for gene regulation. For example, in Candida albicans, Ume6 is an important activator of genes required for hypha formation and undergoes Skp-cullin-F box (SCF)-dependent degradation in response to oxygen or other conditions that inhibit hyphal growth (Citation55, Citation56). SCF is required for sporulation in S. cerevisiae, but early gene expression is not affected by Cdc53 depletion or Cdc4 inactivation (Citation57).
Repressor Rpd3 remains associated with Ume6 when early genes are activated.
It was expected that the lysine deacetylase Rpd3 would be associated with Ume6 at early meiosis-specific gene promoters during vegetative growth, but it was surprising to find that it remained associated with these promoters while those genes were induced. We did not probe for the presence of Sin3 in these complexes, but Sin3’s abundance does not change during sporulation and Ume6-Sin3 complexes have been detected during early and middle phases of sporulation (Citation47, Citation58). Sin3 physically interacts with Ume6 through contact with specific residues in a Sin3 Pah domain (Citation51). In contrast, there is no evidence for direct binding of Rpd3 to Ume6. Rpd3 is resident in multiple chromatin-bound complexes in cells (Citation46). Indeed, interaction between Rpd3, Sin3, and other subunits of the Ume6 complex may be critical to the recruitment and retention of Rpd3 on early meiosis-specific gene promoters (Citation15, Citation59). Distinct from the findings of Inai et al. (Citation47) and those reported here, Pnueli et al. observed transient displacement of Rpd3 from the IME2 promoter at the onset of sporulation (Citation23). Those investigators monitored the Rpd3-13×MYC occupancy of IME2 carried on a 2μ plasmid, which may have given a different result from our monitoring of occupancy of the endogenous IME2 and SPO13 promoters by an endogenously tagged Rpd3-3×HA.
We were able to identify Ume6-Rpd3 and Ume6-Ime1 complexes bound to early meiosis-specific gene promoters during the early phase of sporulation, and there is evidence that Ime1 and Rpd3 co-reside in at least a subset of those complexes. A simple interpretation of these data would suggest that Rpd3 remains bound to Ume6 following the recruitment of Ime1. However, the possibility that Rpd3 is transiently displaced but not easily detectable within the cell population at the level of resolution allowed by our synchronization protocol cannot be precluded. A caveat to this aspect of our investigation is that Rpd3 binds to several complexes, not only Ume6, and has been shown to spread over some chromosomal regions (Citation46). It is possible that Rpd3 does dissociate from Ume6 but remains in close proximity, associated with other chromatin complexes, and our procedure does not have sufficient resolution to distinguish between them.
It is unclear exactly how the repressive effects of Sin3-Rpd3 are bypassed when Ime1 joins the Ume6 complex during the sporulation process. It may be that binding to Ime1 leads to rearrangement of the three-dimensional structure of the complex, rendering Rpd3 unable to deacetylate surrounding chromatin or relevant proteins within the complex. Alternatively, Ime1 or some other meiosis-specific factor might inhibit Rpd3 activity. Rpd3L complexes containing Ume6 and Rpd3 display reduced histone deacetylase activity when cells are cultured in acetate medium (Citation60). The repressive activity of Rpd3 is dependent on the activator it competes with and the degree of activation (Citation61). Thus, it may simply be that Ime1 is a sufficiently strong activator to overpower the repressors while it is bound to Ume6.
Rpd3-Sin3 in conjunction with Isw2 promotes an inhibitory chromatin structure that masks access to the TATA sequence of meiosis-specific genes IME2 and HOP1 (Citation62). This inhibitory chromatin structure is overcome by recruitment of Ime1, which promotes binding of the TATA box binding protein (TBP) encoded by SPT15 to the TATA sequence of HOP1 (Citation62). Recruitment of TBP to the IME2 promoter by Ime1 has not been specifically demonstrated, but TBP binding is promoted by other transcriptional trans activators (Citation39). A variety of structural and genetic data support a model whereby Tra1 as a component of the SAGA complex binds directly to trans activators, bringing Gcn5 and TBP to specific promoters to induce transcriptional activation (Citation63).
The expression of early meiosis-specific genes is dependent not only on Ime1 but also on Gcn5. The chromatin surrounding early gene promoters displays an increase in acetylated histones at the onset of sporulation that is dependent upon Gcn5, and the early genes are not induced in gcn5 mutants (Citation40). The relationship between Ime1 and Gcn5 is not entirely clear. We observe that Gcn5’s association with the IME2 promoter is largely though not entirely dependent upon Ime1, suggesting that Tra1 and the SAGA complex may bind Ime1 as they do with the activators Gal4 and Gcn4 (Citation38, Citation49). This observation does not preclude the possibility that other transcriptional activating complexes also participate in this process. Gcn5 is known to be a component of the SAGA, SLIK, and ADA complexes (Citation64, Citation65). This interpretation is subject to several caveats, as we have not demonstrated direct interaction between Tra1 and Ime1 or any component of the Ume6 complex. Furthermore, the Ume6-Isw2 complex can in some cases promote chromatin loop formation, which could potentially allow cross-linking with proteins associated with the promoters of genes that are not meiosis-specific and cloud the interpretation of results at specific promoters (Citation66).
Ime1 may not be strictly required for histone acetylation surrounding the IME2 promoter, as some increase in histone H3 acetylation has been reported in ime1 mutants that are incubated in sporulation medium (Citation47). In gcn5 mutants, there is some evidence that Ime1 does not bind stably to promoter chromatin, suggesting that acetylation of histones or some component of the Ume6 complex is required for stable Ime1 interaction (Citation47). It is clear that Gcn5 is required for more than reversal of Rpd3-dependent histone deacetylation, since Gcn5 is required for sporulation even in the absence of Rpd3 (Citation40). In addition to histones, Gcn5 can acetylate Ume6 (Citation41), and it is possible that acetylation of Ume6 is required for Ime1 to associate stably with the complex. This scenario might explain the failure to induce early genes in a gcn5 mutant and the weak induction of early genes in a strain harboring a mutated Ume6 that cannot be acetylated (Citation40, Citation41).
The data reported here support a role for Ume6 as a binding platform for both activators and repressors of early meiosis-specific genes. Stable association of Ume6 with chromatin throughout the sporulation process is important for the activation of early genes and may be important for the ability to reimpose repression as cells progress into the middle and late phases of sporulation. Additionally, Ume6 plays an important role in spore germination (Citation67). Thus, retaining the complex on chromatin and regulating meiosis-specific expression through the stability and posttranslational regulation of the activator Ime1 may be more economical for cells than dissociating or degrading the DNA binding complex, since this would require resynthesis of Ume6 and reassembly of the repressive complex during the late phase of sporulation when cells do not have access to external sources of nitrogen.
MATERIALS AND METHODS
Strains and plasmids.
The Saccharomyces cerevisiae strains used in this study and their relevant genotypes are listed in . Strain EGY48 was provided by Erica Golemis, and its construction has been previously described (Citation68). Strain DSY1725 was derived from EGY48 by transformation. Other strains used throughout this work were derived from the SK1 genetic background (Citation69). Unless otherwise indicated, strains in the SK1 background were derived from DSY1030 MATa ho::LYS2 lys2 ura3 leu2::hisG trp1::hisG arg4-bgl his4B and DSY1031 MATα ho::LYS2 lys2 ura3 leu2::hisG trp1::hisG arg4-nsp his4X. The SK1 diploid strain A5128, generously provided by Angelika Amon, has the endogenous CDC20 promoter replaced by the mitosis-specific CLB2 promoter and includes 3 copies of the HA epitope at the amino terminus of CDC20 as described previously (Citation26). S. cerevisiae strains were routinely maintained on YEPD rich medium (Citation70). Sporulation experiments were performed as previously described (Citation71). Single colonies of each diploid strain were selected from a YEP-glycerol agar plate to ensure mitochondrial function prior to overnight propagation in YEPD medium. Cultures were diluted 1:100 into YEP medium supplemented with 1% potassium acetate (YPKAc) for overnight growth. Cultures were then collected by centrifugation, washed with water, and resuspended in sporulation medium (SPM) (1% KAc, 0.02% raffinose) to initiate sporulation.
TABLE 2 Saccharomyces cerevisiae strains used in this study
The HA-UME6 allele, generously provided by Aaron Mitchell, has 3 tandem copies of the HA epitope introduced into the Ume6 sequence at amino acid 122 (Citation43). The tagged construct was used to replace the endogenous UME6 gene through a two-step gene replacement procedure (Citation72). The carboxyl-terminal MYC or HA tags were introduced into the endogenous UME6, RPD3, SIN3, IME1, and GCN5 open reading frames by PCR-mediated transformation (Citation73). A 900-bp fragment of the TRA1 promoter was amplified from SK1 genomic DNA with oligonucleotides Tra1p5/Tra1p3 and inserted into BglII-/PacI-cut pFA6a-TRP1-GAL1-3xHA. This was used as a template for amplification with Tra1Up2/Tra1dwn. The PCR product was used to tag TRA1 (Citation72). Deletion of the UME6, IME1, and GCN5 genes was accomplished by PCR-mediated transformation (Citation74). Oligonucleotide DNA sequences used to generate tagged genes and gene deletions are listed in . The modified strains, with the exception of DSY1725 (EGY48 gcn5::natMX4), were subjected to two rounds of backcrossing to the parent DSY1030 to ensure that no unwanted mutations had been acquired during the strain construction.
TABLE 3 Oligonucleotides used in this study
Cytology.
Sporulation frequency was determined by counting asci visualized by light microscopy; 200 cells per culture were counted. Progression through sporulation was monitored by fluorescence microscopy. At each time point, 100 μl of culture was fixed in 70% ethanol for at least 4 h at 4°C. Following rehydration, cells were stained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) and visualized; 200 cells per time point were counted. Cells that contained two or more masses of chromatin stained with DAPI were considered to be post-MI.
Protein analysis.
Samples of culture were collected at the indicated time points and protein extracts prepared by bead beating in 20% trichloroacetic acid (TCA) as described previously (Citation75). The cells were pelleted by centrifugation for 1 min at 10,000 × g. The medium was removed, and cell pellets were resuspended in 500 μl of 20% TCA and incubated for 2 min on ice. Cell samples were pelleted by centrifugation, the TCA removed, and the cell pellet resuspended in 200 μl of 20% TCA. Glass beads (0.5 mm) were added to each sample, and the cells were disrupted by vortexing 4 times for 1 min each at full speed and incubating for 1 min on ice between bursts of vortexing. Four hundred microliters of 5% TCA was then added to each sample for a final mixture of 10% TCA. The bottom of each tube was then punctured with a 25-gauge needle and each tube placed into a fresh tube before centrifuging for 3 min at 3,000 × g to separate the cell extract from the beads. The cell extracts were then subjected to centrifugation for 10 min at 14,000 × g. The TCA was removed, and the pelleted extract was resuspended in 1 ml of ice-cold acetone. This was again subjected to centrifugation for 10 min at 14,000 × g. The acetone was removed, and the pellet resuspended in 100 μl of 4% SDS, 20% glycerol, 0.02% bromophenol blue, 100 mM Tris-HCl (pH 6.8), 20 μl of 1 M dithiothreitol (DTT), 30 μl of 1 M Tris base, 50 μl H2O. Samples were stored at −80°C prior to analysis by gel electrophoresis. An alternative protein extraction procedure was performed as described previously (Citation45). The cell pellets taken at each time point were resuspended in 1 ml of ice-cold water, and then 150 μl of 1.85 N NaOH, 7.5% 2-mercaptoethanol was added. This mixture was incubated on ice for 10 min prior to the addition of 150 μl of 55% TCA. Following a 10-min incubation on ice, the extract was centrifuged for 10 min at 14,000 × g. The supernatant was removed, and the pellet was resuspended in 100 μl HU buffer (8 M urea, 5% SDS, 200 mM Tris-HCl [pH 6.8], 1 mM EDTA, 0.1% bromophenol blue, DTT added to a final concentration of 1.5% immediately before use)/2 × 107 cells. Protein extracts were prepared under nondenaturing conditions by resuspending cell pellets in 50 mM Tris-HCl (pH 7.4), 300 mM NaCl, 0.5% NP-40, 10% glycerol (pH 8.0), 20 μg/ml each of pepstatin, leupeptin, and aprotinin, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM sodium fluoride, 60 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA. Glass beads (0.5 mm) were added to each sample, and the cells were disrupted by vortexing 4 times for 1 min each time at full speed and incubating for 1 min on ice between bursts of vortexing. The extracts were subjected to centrifugation for 10 min at 14,000 × g, and the soluble portion was mixed with an equal volume of 4% SDS, 20% glycerol, 200 mM DTT, 0.02% bromophenol blue, 100 mM Tris-HCl, pH 6.8.
Ume6 protein samples were separated by gel electrophoresis using 15-cm 8% SDS–polyacrylamide gels. Proteins were transferred to polyvinylidene fluoride (PVDF) membranes (ImmobilonP) using semidry transfer (48 mM Tris, 39 mM glycine 0.037% SDS, 20% methanol) for 90 min at 200 mA. The membranes were subsequently cut at the 55-kDa marker and separated. The upper half of each membrane was probed for Ume6 by incubating with antibody recognizing the HA or MYC epitope tag or Ume6 as indicated. The lower half of each membrane was probed for Cdc28 by incubation with an anti-PSTAIRE antibody recognizing the amino acid sequence EGVPSTAIREISLLKE in Cdc28. The antibodies used were as follows: anti-HA mouse monoclonal antibody HA.11 (clone 16B12, MMS-101R; Covance), 3 mg/ml ascites fluid used at 1:10,000 dilution; anti-MYC mouse monoclonal antibody 9E10 (MMS-150R; Covance), 4.1 mg/ml ascites fluid used at 1:10,000 dilution; affinity-purified anti-Ume6 chicken polyclonal antibody (GW22454A, batch number 039K2073V; Sigma-Aldrich), 1 mg/ml used at 1:500 dilution; and anti-PSTAIRE mouse IgG1 monoclonal antibody (P7962, lot number 010M4766; Sigma-Aldrich), 7.2 mg/ml ascites fluid used at 1:10,000 dilution. Primary anti-HA and anti-MYC antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (12-349; Millipore Sigma). Primary anti-Ume6 antibody was detected with HRP-conjugated anti-chicken IgY (A9046, batch number 113M4824). For the quantitative Western blot analysis whose results are shown in , the same primary antibodies were employed; these were detected with the following IRDye-coupled secondary antibodies as appropriate: IRDye 680RD donkey anti-chicken IgY 926-68075 (Ume6) or IRDye 680RD goat anti-mouse IgG 926-68070 (Ume6-MYC) (LI-COR Biosciences). The Odyssey FC infrared imaging system scanner (LI-COR Biosciences) was used to visualize the decorated proteins in the 700-nm and 800-nm channels. Quantification of the band intensities was performed using ImageJ, and the relative amount of Ume6 or Ume6-MYC was normalized to the amount of the loading control Cdc28. Sin3-MYC samples were separated in 6.4-cm 6% SDS–polyacrylamide gels. Rpd3-HA samples were separated in 6.4-cm 10% SDS–polyacrylamide gels. Western blot detection was performed using the appropriate primary antibody and HRP-conjugated secondary antibodies as described above. Duplicate samples were electrophoresed in 10% SDS–polyacrylamide gels for Western blot detection of Cdc28.
Analysis of gene expression.
Samples of total RNA collected from actively proliferating cells and at intervals following the induction of sporulation were analyzed by Northern blotting using 32P-labeled PCR-generated probe fragments as previously described (Citation76). Repression of the PIME2-LacZ reporter and activation of the LEXAop-LacZ reporter was monitored by p-nitrophenyl-α-d-galactopyranoside (ONPG) assay as described previously (Citation77). Six independent cultures of each strain actively proliferating in SD-ura medium (Citation70) were assayed. Statistical significance was determined by the use of an unpaired t test using the GraphPad t test calculator.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed based on previously described protocols (Citation78). Samples of 2 × 108 cells, either mitotically proliferating or sporulating, were collected by centrifugation and washed with phosphate-buffered saline (PBS), followed by cross-linking for 45 min by the addition of 10 mM dimethyl adipimidate (DMA) in PBS plus 0.25% dimethyl sulfoxide (DMSO). The DMA was subsequently washed out, and the cells were subjected to further cross-linking in the presence of 1% formaldehyde for 60 min. The cross-linking reaction was quenched by the addition of glycine to a final concentration of 125 mM. The samples were washed four times in 4 volumes of PBS. Cell pellets were lysed by beating with 0.5-mm glass beads in FA lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 20 μg/ml each of aprotinin, leupeptin, pepstatin, 1 mM PMSF), and chromatin was sheared by sonicating 10 times for 15 s each time with a 15 s rest on ice between bursts to achieve an average size of 300 to 400 bp. Following centrifugation for 10 min at 13,000 × g, 1/10 of each lysate was reserved as a whole-cell extract (WCE) control and the remaining supernatant fraction was precleared by the addition of protein G-Sepharose beads that had been blocked with salmon sperm DNA. The target proteins were immunoprecipitated with either 100 μg anti-MYC antibodies (9E10, MMS150R; Covance) or 100 μg anti-HA antibodies (HA.11, MMS-101; Covance) in ascites fluid and salmon sperm-blocked protein G-Sepharose beads. The antibody-bead complexes were washed once with FA lysis buffer, once with FA lysis buffer supplemented with NaCl to a final concentration of 500 mM, once with ChIP wash buffer (10 mM Tris-HCl at pH 8.0, 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% [wt/vol] sodium deoxycholate), and twice with 10 mM Tris at pH 8.0, 1 mM EDTA, for 5 min each time. Protein-DNA complexes were eluted using ChIP elution buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 1% SDS). Cross-linking was reversed by incubation at 95°C for 10 min and 65°C overnight followed by proteinase K digestion at 42°C for 3 h. The WCE samples were prepared similarly. DNA was subsequently isolated using PCR purification spin columns (Qiagen). For ChIP re-ChIP experiments, the initial immunoprecipitate was incubated in ChIP elution buffer for 30 min, followed by centrifugation. The supernatant was diluted to 1 ml with FA buffer and was reprecipitated with the second antibody. The DNA samples recovered were subjected to 15 cycles of PCR with 32P-end-labeled oligonucleotide pairs (2 pmol of each oligonucleotide). Oligonucleotide sequences used for PCR analysis are listed in . The SPO13 oligonucleotide pair amplifies a DNA fragment, including the URS1 at −96. The SPO13 pair amplifies a DNA region encompassing URS1 sequences at −456 and −551. The TELVI oligonucleotide pair amplifies a fragment from the right arm of the chromosome VI telomere that has no URS1 sequence. The PCR products were separated on 6% polyacrylamide gels, and a Molecular Dynamics STORM 840 phosphor imager was used to scan and quantitate the radioactive signals. To determine relative enrichment, the PCR signals are reported as the ratios of immunoprecipitated DNA/input DNA. All ChIP analyses were performed in triplicate.
Chromatin immunoprecipitation experiments for Gcn5 and Tra1 were analyzed by qPCR. Chromatin immunoprecipitation was performed as described above, but 2% of the cell lysate was reserved as the input control. The remaining extract was split, and half was subjected to immunoprecipitation with anti-MYC or anti-HA antibodies in ascites fluid (10 μg) and protein G-Sepharose while half received no antibodies (no antibody control). The input and immunoprecipitated DNA were subjected to qPCR analysis as follows: total volume of 10 μl containing 10 pmol of forward and reverse primers (IME2SC5/IME2SC3 or TEL6B/TEL6C), 5.0 μl 2× iQ SYBR green supermix (Bio-Rad), and 2 μl of the sample as the template. Amplifications were performed using the Rotor-Gene Q real-time PCR cycler. No-template controls were included in each reaction mixture, and all samples were tested in technical triplicates. The amplification efficiency of all primer pairs was tested prior to use with serial dilutions of sonicated chromosomal DNA to confirm that amplification efficiency was greater than 97%. Enrichment was calculated, as previously described (Citation79), by determining the percentage of input DNA recovered in each IME2 promoter-specific immunoprecipitation minus no-antibody control divided by input DNA. The values recovered from the IME2 promoter immunoprecipitations were then divided by the percentages of input DNA recovered from the TELVI immunoprecipitations. Data shown reflect three independent immunoprecipitations and three technical replicates of each. Error bars indicate standard deviations.
ACKNOWLEDGMENTS
We thank Aaron Mitchell for the HA-tagged UME6, Angelika Amon for PCLB2-CDC20 strain A5128, and Erica Golemis for yeast strain EGY48. We thank Bonnie McNeil for helpful advice regarding performing the qPCR analysis.
Funding for this project was provided by Natural Sciences and Engineering Research Council of Canada (NSERC) discovery grant number RGPIN-2015-03673 to D.T.S.
The project was conceived by D.T.S. The strain construction and experimental work were performed by S.A.R., S.V., M.T.I., B.G., and D.T.S. The manuscript was written and revised by D.T.S.
REFERENCES
- Silveira MAD, Bilodeau S. 2018. Defining the transcriptional ecosystem. Mol Cell 72:920–924. https://doi.org/10.1016/j.molcel.2018.11.022.
- Neiman AM. 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189:737–765. https://doi.org/10.1534/genetics.111.127126.
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699–705. https://doi.org/10.1126/science.282.5389.699.
- Chu S, Herskowitz I. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell 1:685–696. https://doi.org/10.1016/S1097-2765(00)80068-4.
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. 2000. The core meiotic transcriptome in budding yeasts. Nat Genet 26:415–423. https://doi.org/10.1038/82539.
- Chia M, van Werven FJ. 2016. Temporal expression of a master regulator drives synchronous sporulation in budding yeast. G3 (Bethesda) 6:3553–3560. https://doi.org/10.1534/g3.116.034983.
- Kassir Y, Granot D, Simchen G. 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853–862. https://doi.org/10.1016/0092-8674(88)90427-8.
- Smith HE, Su SS, Neigeborn L, Driscoll SE, Mitchell AP. 1990. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol 10:6103–6113. https://doi.org/10.1128/MCB.10.12.6103.
- Mandel S, Robzyk K, Kassir Y. 1994. IME1 gene encodes a transcription factor which is required to induce meiosis in Saccharomyces cerevisiae. Dev Genet 15:139–147. https://doi.org/10.1002/dvg.1020150204.
- Williams RM, Primig M, Washburn BK, Winzeler EA, Bellis M, Sarrauste de Menthiere C, Davis RW, Esposito RE. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc Natl Acad Sci U S A 99:13431–13436. https://doi.org/10.1073/pnas.202495299.
- Anderson SF, Steber CM, Esposito RE, Coleman JE. 1995. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci 4:1832–1843. https://doi.org/10.1002/pro.5560040918.
- Kadosh D, Struhl K. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365–371. https://doi.org/10.1016/S0092-8674(00)80217-2.
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol 21:6450–6460. https://doi.org/10.1128/MCB.21.19.6450-6460.2001.
- Kadosh D, Struhl K. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol 18:5121–5127. https://doi.org/10.1128/MCB.18.9.5121.
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831–835. https://doi.org/10.1038/33952.
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423–433. https://doi.org/10.1016/S0092-8674(00)00134-3.
- Bowdish KS, Mitchell AP. 1993. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol Cell Biol 13:2172–2181. https://doi.org/10.1128/MCB.13.4.2172.
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev 8:796–810. https://doi.org/10.1101/gad.8.7.796.
- Vidan S, Mitchell AP. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol 17:2688–2697. https://doi.org/10.1128/MCB.17.5.2688.
- Colomina N, Gari E, Gallego C, Herrero E, Aldea M. 1999. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J 18:320–329. https://doi.org/10.1093/emboj/18.2.320.
- Smith HE, Driscoll SE, Sia RA, Yuan HE, Mitchell AP. 1993. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics 133:775–784. https://doi.org/10.1093/genetics/133.4.775.
- Bowdish KS, Yuan HE, Mitchell AP. 1995. Positive control of yeast meiotic genes by the negative regulator UME6. Mol Cell Biol 15:2955–2961. https://doi.org/10.1128/MCB.15.6.2955.
- Pnueli L, Edry I, Cohen M, Kassir Y. 2004. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol Cell Biol 24:5197–5208. https://doi.org/10.1128/MCB.24.12.5197-5208.2004.
- Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol 16:2518–2526. https://doi.org/10.1128/MCB.16.5.2518.
- Mallory MJ, Cooper KF, Strich R. 2007. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol Cell 27:951–961. https://doi.org/10.1016/j.molcel.2007.08.019.
- Lee BH, Amon A. 2003. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300:482–486. https://doi.org/10.1126/science.1081846.
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. 2005. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell 120:773–788. https://doi.org/10.1016/j.cell.2005.01.032.
- Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, Markova Z, Zagoriy I, Novak B, Zachariae W. 2012. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 151:603–618. https://doi.org/10.1016/j.cell.2012.08.044.
- Tan GS, Lewandowski R, Mallory MJ, Strich R, Cooper KF. 2013. Mutually dependent degradation of Ama1p and Cdc20p terminates APC/C ubiquitin ligase activity at the completion of meiotic development in yeast. Cell Div 8:9. https://doi.org/10.1186/1747-1028-8-9.
- Marston AL, Tham WH, Shah H, Amon A. 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303:1367–1370. https://doi.org/10.1126/science.1094220.
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci U S A 97:14548–14553. https://doi.org/10.1073/pnas.250351297.
- Diamond AE, Park JS, Inoue I, Tachikawa H, Neiman AM. 2009. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol Biol Cell 20:134–145. https://doi.org/10.1091/mbc.e08-06-0615.
- Penkner AM, Prinz S, Ferscha S, Klein F. 2005. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell 120:789–801. https://doi.org/10.1016/j.cell.2005.01.017.
- Tan GS, Magurno J, Cooper KF. 2011. Ama1p-activated anaphase-promoting complex regulates the destruction of Cdc20p during meiosis II. Mol Biol Cell 22:315–326. https://doi.org/10.1091/mbc.e10-04-0360.
- Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell 3:1464–1475. https://doi.org/10.1128/EC.3.6.1464-1475.2004.
- Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, Inoki K, Klionsky DJ. 2012. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A 109:11206–11210. https://doi.org/10.1073/pnas.1200313109.
- Wen FP, Guo YS, Hu Y, Liu WX, Wang Q, Wang YT, Yu HY, Tang CM, Yang J, Zhou T, Xie ZP, Sha JH, Guo X, Li W. 2016. Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy 12:671–688. https://doi.org/10.1080/15548627.2016.1149659.
- Bhaumik SR, Green MR. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15:1935–1945. https://doi.org/10.1101/gad.911401.
- Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol 25:3461–3474. https://doi.org/10.1128/MCB.25.9.3461-3474.2005.
- Burgess SM, Ajimura M, Kleckner N. 1999. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Natl Acad Sci U S A 96:6835–6840. https://doi.org/10.1073/pnas.96.12.6835.
- Mallory MJ, Law MJ, Sterner DE, Berger SL, Strich R. 2012. Gcn5p-dependent acetylation induces degradation of the meiotic transcriptional repressor Ume6p. Mol Biol Cell 23:1609–1617. https://doi.org/10.1091/mbc.e11-06-0536.
- Steber CM, Esposito RE. 1995. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc Natl Acad Sci U S A 92:12490–12494. https://doi.org/10.1073/pnas.92.26.12490.
- Xiao Y, Mitchell AP. 2000. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol Cell Biol 20:5447–5453. https://doi.org/10.1128/MCB.20.15.5447-5453.2000.
- Bryant JM, Govin J, Zhang L, Donahue G, Pugh BF, Berger SL. 2012. The linker histone plays a dual role during gametogenesis in Saccharomyces cerevisiae. Mol Cell Biol 32:2771–2783. https://doi.org/10.1128/MCB.00282-12.
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972. https://doi.org/10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W.
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet 31:248–254. https://doi.org/10.1038/ng907.
- Inai T, Yukawa M, Tsuchiya E. 2007. Interplay between chromatin and trans-acting factors on the IME2 promoter upon induction of the gene at the onset of meiosis. Mol Cell Biol 27:1254–1263. https://doi.org/10.1128/MCB.01661-06.
- Sharov G, Voltz K, Durand A, Kolesnikova O, Papai G, Myasnikov AG, Dejaegere A, Ben Shem A, Schultz P. 2017. Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA. Nat Commun 8:1556. https://doi.org/10.1038/s41467-017-01564-7.
- Lin L, Chamberlain L, Zhu LJ, Green MR. 2012. Analysis of Gal4-directed transcription activation using Tra1 mutants selectively defective for interaction with Gal4. Proc Natl Acad Sci U S A 109:1997–2002. https://doi.org/10.1073/pnas.1116340109.
- Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, Moorefield B, Pacheco D, Hahn S. 2010. Mechanism of mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol Cell Biol 30:2376–2390. https://doi.org/10.1128/MCB.01046-09.
- Washburn BK, Esposito RE. 2001. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol Cell Biol 21:2057–2069. https://doi.org/10.1128/MCB.21.6.2057-2069.2001.
- Horvath A, Riezman H. 1994. Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10:1305–1310. https://doi.org/10.1002/yea.320101007.
- Byers B, Goetsch L. 1982. Reversible pachytene arrest of Saccharomyces cerevisiae at elevated temperature. Mol Gen Genet 187:47–53. https://doi.org/10.1007/BF00384382.
- Lardenois A, Becker E, Walther T, Law MJ, Xie B, Demougin P, Strich R, Primig M. 2015. Global alterations of the transcriptional landscape during yeast growth and development in the absence of Ume6-dependent chromatin modification. Mol Genet Genomics 290:2031–2046. https://doi.org/10.1007/s00438-015-1051-5.
- Mendelsohn S, Pinsky M, Weissman Z, Kornitzer D. 2017. Regulation of the Candida albicans hypha-inducing transcription factor Ume6 by the CDK1 cyclins Cln3 and Hgc1. mSphere 2:e00248-16. https://doi.org/10.1128/mSphere.00248-16.
- Lu Y, Su C, Ray S, Yuan Y, Liu H. 2019. Signaling through the Ptc2-Ssn3 axis governs sustained hyphal development of Candida albicans by reducing Ume6 phosphorylation and degradation. mBio 10:e02320-18. https://doi.org/10.1128/mBio.02320-18.
- Zhu Z, Bani Ismail M, Shinohara M, Shinohara A. 2021. SCFCdc4 ubiquitin ligase regulates synaptonemal complex formation during meiosis. Life Sci Alliance 4:e202000933. https://doi.org/10.26508/lsa.202000933.
- Lamb TM, Mitchell AP. 2001. Coupling of Saccharomyces cerevisiae early meiotic gene expression to DNA replication depends upon RPD3 and SIN3. Genetics 157:545–556. https://doi.org/10.1093/genetics/157.2.545.
- Mallory MJ, Strich R. 2003. Ume1p represses meiotic gene transcription in Saccharomyces cerevisiae through interaction with the histone deacetylase Rpd3p. J Biol Chem 278:44727–44734. https://doi.org/10.1074/jbc.M308632200.
- Yukawa M, Yo K, Hasegawa H, Ueno M, Tsuchiya E. 2009. The Rpd3/HDAC complex is present at the URS1 cis-element with hyperacetylated histone H3. Biosci Biotechnol Biochem 73:378–384. https://doi.org/10.1271/bbb.80621.
- Deckert J, Struhl K. 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol Cell Biol 22:6458–6470. https://doi.org/10.1128/MCB.22.18.6458-6470.2002.
- Shimizu M, Takahashi K, Lamb TM, Shindo H, Mitchell AP. 2003. Yeast Ume6p repressor permits activator binding but restricts TBP binding at the HOP1 promoter. Nucleic Acids Res 31:3033–3037. https://doi.org/10.1093/nar/gkg425.
- Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, Ben-Shem A. 2020. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577:711–716. https://doi.org/10.1038/s41586-020-1944-2.
- Wang L, Liu L, Berger SL. 1998. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev 12:640–653. https://doi.org/10.1101/gad.12.5.640.
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, III, Grant PA. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol 22:8774–8786. https://doi.org/10.1128/MCB.22.24.8774-8786.2002.
- Yadon AN, Singh BN, Hampsey M, Tsukiyama T. 2013. DNA looping facilitates targeting of a chromatin remodeling enzyme. Mol Cell 50:93–103. https://doi.org/10.1016/j.molcel.2013.02.005.
- Strich R, Khakhina S, Mallory MJ. 2011. Ume6p is required for germination and early colony development of yeast ascospores. FEMS Yeast Res 11:104–113. https://doi.org/10.1111/j.1567-1364.2010.00696.x.
- Estojak J, Brent R, Golemis E. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol 15:5820–5829. https://doi.org/10.1128/MCB.15.10.5820.
- Kane SM, Roth R. 1974. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol 118:8–14. https://doi.org/10.1128/JB.118.1.8-14.1974.
- Sherman F. 2002. Getting started with yeast. Methods Enzymol 350:3–41. https://doi.org/10.1016/s0076-6879(02)50954-x.
- Stuart D, Wittenberg C. 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev 12:2698–2710. https://doi.org/10.1101/gad.12.17.2698.
- Scherer S, Davis RW. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A 76:4951–4955. https://doi.org/10.1073/pnas.76.10.4951.
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. https://doi.org/10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U.
- Wach A, Brachat A, Pohlmann R, Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808. https://doi.org/10.1002/yea.320101310.
- Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. 1994. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol 14:923–933. https://doi.org/10.1128/MCB.14.2.923.
- Raithatha SA, Stuart DT. 2005. Meiosis-specific regulation of the Saccharomyces cerevisiae S-phase cyclin CLB5 is dependent on MluI cell cycle box (MCB) elements in its promoter but is independent of MCB-binding factor activity. Genetics 169:1329–1342. https://doi.org/10.1534/genetics.104.036103.
- Rupp S. 2002. LacZ assays in yeast. Methods Enzymol 350:112–131. https://doi.org/10.1016/s0076-6879(02)50959-9.
- Kurdistani SK, Grunstein M. 2003. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods 31:90–95. https://doi.org/10.1016/S1046-2023(03)00092-6.
- Ahmed NT, Bungard D, Shin ME, Moore M, Winter E. 2009. The Ime2 protein kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters. Mol Cell Biol 29:4352–4362. https://doi.org/10.1128/MCB.00305-09.
- Gyuris J, Golemis E, Chertkov H, Brent R. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791–803. https://doi.org/10.1016/0092-8674(93)90498-F.
