Abstract
T-helper 1 polarization in patients with primary immune thrombocytopenia (ITP) is well documented. However, the genetic contribution to this imbalance remains unclear. To address this question, we selected six candidate single nucleotide polymorphisms within cytokine or cytokine receptor genes for association testing among Caucasian adults. Patients from the United Kingdom Adult ITP Registry were gender-matched (1∶3) with healthy controls from the Wellcome Trust Case Control Consortium. Variants IL10 −819 c>t, TNFA −308 g>a, TGFB1 −509 c>t, IL1A −889 c>t, IL10 −592 c>t, and IL4R q576r were measured in cases and retrieved for controls from the European Genome-phenome Archive. Associations were evaluated using logistic regression models. In total, 206 patients with primary ITP were matched with 618 controls. A significant per allele odds ratio of 1·34 (95% confidence interval, 1·03–1·75; P = 0·03) was observed for TNFA −308 g>a, implicating an increased disease susceptibility among Caucasian carriers of the rare allele.
Introduction
Primary immune thrombocytopenia (ITP) is an acquired autoimmune disease characterized by a reduced platelet count in the absence of a known cause. It is hypothesized to result from a complex interplay of genetic and environmental factors leading to the development of autoantibodies against platelet and megakaryocyte glycoproteins.Citation1 The phenotype of primary ITP exhibits considerable variability in adults. Although one-fourth of patients are asymptomatic at diagnosis (i.e. diagnosed incidentally),Citation2 clinically relevant morbidity such as major haemorrhage is not uncommon.Citation3
Table 1. Observed genotype frequencies and SNP–disease, Helicobacter pylori infection, and severity association results
The traditional T-helper 1 (Th1)/Th2 model for the classification of Th cells has been used to evaluate autoimmune diseases. Under this paradigm, homeostasis is maintained through the balance of cytokines secreted by Th1 [e.g. interferon-gamma, interleukin-2 (IL-2), tumour necrosis factor (TNF)-alpha, and TNF-beta1] and Th2 (e.g. IL-4, IL-5, and IL-6) cells.Citation4 While the former class of Th1 cells promotes pro-inflammatory, cell-mediated, and complement-fixing IgG isotype (IgG3 and IgG1) responses effective in the combat of intracellular pathogens, the latter elicits immediate-type hypersensitivity, which augments humoral defense against extracellular pathogens.Citation5 Crucially, Th1 and Th2 cell responses further demonstrate down regulatory effects on each other.Citation5
Past investigations have demonstrated Th1 polarization in primary ITP. Wang et al.Citation6 reported a significantly higher Th1/Th2 ratio in adult patients with chronic primary ITP versus healthy controls while Panitsas et al.Citation7 documented an inverse correlation of the Th1/Th2 ratio with platelet count. Moreover, in children with chronic or acute primary ITP, Semple et al.Citation8 failed to detect IL-4 or IL-6, prototypical Th2 cytokines.
The genetic contribution to this imbalance is unclear. Although common susceptibility loci for autoimmune diseases have been reported,Citation9 few studies have investigated the role of genetic polymorphisms in primary ITP among adults, with none having evaluating the role of single nucleotide polymorphisms (SNPs) within cytokine or cytokine receptor genes in an entirely Caucasian population. The primary objective of our study, therefore, was to test for associations of candidate SNPs in cytokine or cytokine receptor genes with primary ITP among Caucasian adults.
Methods
Study design and selection of candidate SNPs
A case–control design was adopted and six SNPs within cytokine or cytokine receptor genes were selected on the basis of their association with other autoimmune diseases: IL10 −819 c>t [rs1800872; rheumatoid arthritis (RA)],Citation10 TNFA −308 g>a (rs1800629; RA),Citation11 TGFB1 −509 c>t (rs1800469; chronic idiopathic neutropenia),Citation12 IL1A −889 c>t (rs1800587; systemic lupus erythematosus),Citation13 IL10 −592 c>t (rs1800871; RA),Citation10 and IL4R q576r (rs1801275; atopic disorders).Citation14
Study subjects
The study was conducted under the auspices of the United Kingdom (UK) Adult ITP Registry, and informed consent was obtained from all patients upon entry. Primary ITP cases comprised Caucasian adults (>16 years) from the UK or New York Presbyterian Hospital who had enrolled in the Registry prior to 1 January 2006. Cases were gender-matched (1∶3) with healthy controls from the Wellcome Trust Case Control Consortium subset of the 1958 British Birth Cohort, which was restricted to the 97% of the cohort identified as Caucasian.
Control data
Encrypted genome-wide data (Illumina 1·2M chip; Essex, UK) on controls were secured online from the European Genome-phenome Archive (www.ebi.ac.uk/ega), with SNP-specific data extraction performed using GTOOL v0·4·1 (Oxford, UK).
Laboratory methods
Blood samples for cases were stored at −80°C and thawed in batches prior to DNA extraction. DNA was isolated from sample aliquots using a Qiagen QIAamp DNA Blood Mini Kit (Hilden, Germany). The quality and quantity of extracted DNA were assessed using a NanoDrop ND-1000 spectrophotometer (Wilmington, DE, USA).
Assays were obtained from Applied Biosystems (Foster City, CA, USA). Reactions were prepared using a TaqMan Fast Universal Master Mix (2×), SNP Genotyping Assay Mix (40×), DNase-free water, and genomic DNA (10 ng). PCR amplification was completed using an ABI Prism 7900HT Sequence Detection System under the following conditions: 10 minutes at 95°C followed by 45 cycles at 92°C for 15 seconds and at 60°C for 1 minute.
Clinical data
Helicobacter pylori infection status, disease severity and chronicity, and responses to splenectomy and to isolated first courses of prednisolone, IVIg, anti-D, and rituximab were recorded for patients for whom data were available. A platelet count of less than 10×109/l at presentation served as a surrogate marker for severe disease. Consensus guidelines by the International Working Group on ITPCitation15 were used to define chronicity (>1 year) and complete response (>100×109/l within specified intervals) to treatment.
Statistical analyses
Associations of candidate SNPs with primary ITP were evaluated using logistic regression models of log-additive co-dominance. Results were reported as per rare allele odds ratios (ORs) with 95% confidence intervals (CIs). To avoid underpowered testing, a minimum of 23 cases, calculated using Quanto v1·2·4 (Los Angeles, CA, USA), was required for association analyses with clinical variables. This threshold was formulated using the following specifications: design = unmatched case–control (1∶2), inheritance = log-additive, rare allele frequency = 0·35, baseline risk = 0·20, alpha (two-tailed) = 0·05, 1-beta = 0·80, and an estimated OR = 3·0.
Results and Discussion
In total, 206 (145 females and 61 males) Caucasian adults with primary ITP were gender-matched (1∶3) with 618 (435 females and 183 males) healthy Caucasian controls. The quantity and quality of DNA extracted from case samples were good; the median yield and mean 260/280 absorbance ratio were 175·39 ng/nl (interquartile range: 61·59–407·85 ng/nl) and 1·85±0·28, respectively. As illustrated in both and , logistic regression analysis revealed a significant association of primary ITP with TNFA −308 g>a (OR = 1·34; 95% CI = 1·03–1·75; P = 0·03).
Figure 1. Results of logistic regression modelling of associations between six functional, candidate SNPs and primary using a sample of 206 Caucasian adults with primary ITP gender-matched with 618 healthy Caucasian controls. A statistically significant association was observed between the disease and TNFA −308 g>a.
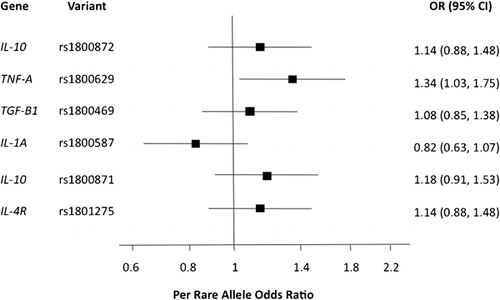
Four previous studies on primary ITP have evaluated TNFA −308 g>a. In a pilot study involving 37 cases and 218 controls, Foster et al.Citation16 reported a significant association (association P = 0·0032; Hardy-Weinberg equilibrium P = 0·02) between the SNP and chronic ITP among children. Investigations by Satoh et al.Citation17 and Suzuki et al.Citation18 were conducted among Japanese cohorts. Although Satoh et al. did not observe a significant association,Citation17 the frequency of the rare allele in the Japanese population is significantly lower than that among Caucasians (HapMap-JPT: 2·3% versus HapMap-CEU: 21·7%). As a result, between 1502 and 3796 Japanese patients with primary ITP would be required to detect an association of similar magnitude as presented in this report. Only 84 adults were included in their study. Suzuki et al., meanwhile, evaluated the association of the SNP with H. pylori infection in patients with primary ITP and not primary ITP itself.Citation18 More recently, Rocha et al.Citation19 failed to uncover a significant association between TNFA −308 g>a and chronic ITP among a Brazilian population (n = 122 adult cases). However, it is unclear what proportion of this cohort was comprised of Caucasians and therefore how many cases would be required to detect the association reported herein.
The principle finding of a borderline significant OR of 1·34 (95% CI, 1·03–1·75; P = 0·03) raises the question as to whether the uncovered association between TNFA −308 g>a and primary ITP among Caucasian adults may constitute a false-positive result. As Wacholder et al.Citation20 commented, the evaluation of such a question should include not only the reported P value, but also the statistical power of the study and the prior probability of a true association. While the detected P value was admittedly moderate, the statistical power of the study was limited and the prior probability of the association was arguably high, suggesting that the finding may constitute a true-positive result. The observed association between TNFA −308 g>a and primary ITP among Caucasian adults is consistent with the latter’s categorization as a Th1-disease. TNF-alpha plays a critical role in the development of pro-inflammatory Th1 responses,Citation21 and the rare allele at −308 has been linked with elevated production of the cytokine. In a Dutch study of patients with inflammatory bowel disease and healthy controls, Bouma et al.Citation22 reported that participants with TNF-E and TNF-C haplotypes secreted the highest and lowest levels of TNF-alpha, respectively. Importantly, these haplotypes differed only at the −308 g>a position (TNF-E: −308 a; TNF-C: −308 g).Citation22 Through in vitro investigations, moreover, Wilson et al.Citation23 have shown the a allele to be a more powerful transcriptional activator than the g allele. As a functional polymorphism, then, TNFA −308 g>a may serve as a marker of pro-inflammatory events.
Clinical data were available for 93 (45·1%) patients with primary ITP, spanning a post-diagnosis follow-up time of 6·0 years (range: 0·4–34·7 years). However, of the variables captured, only H. pylori infection and disease severity met the threshold established for proceeding with analyses, and no significant associations were observed ( and ).
Figure 2. Results of logistic regression modelling of associations between six functional, candidate and both disease severity and H. pylori infection in Caucasian adults with primary ITP. No statistically significant associations were observed.
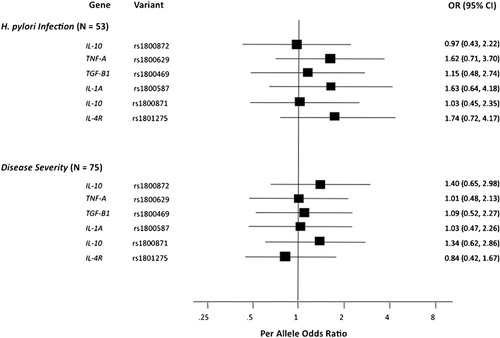
While these findings may reflect true lack of associations, a primary limitation of this study is that the clinical analyses conducted possessed inadequate statistical power. The minimum threshold of cases established for the above association analyses was predicated on the remote possibility of uncovering a pronounced effect (e.g. OR = 3·0). However, SNP associations with these outcomes would likely be less pronounced, were they to exist, and would therefore require a greater sample size to detect. For example, assuming an unmatched case–control (1∶2) design with log-additive inheritance, a rare allele frequency of 0·35, and a baseline risk of 0·20, 155 cases, or patients with severe disease or H. pylori infections, would be required to uncover an OR of 1·5 using significance and power thresholds of 0·05 and 0·80, respectively.
Two further potential limitations of this study should be noted relating to methodological differences in the evaluation of cases and controls. First, the use of different genotyping platforms may have led to spurious differences in SNP frequencies between cases and controls. Second, reliance on self-declared ethnicity for cases may have resulted in a greater admixture, or accidental inclusion of non-Caucasian patients, than among controls, for whom principal component analyses had been performed. Although the possibility that these systematic differences may have biased the results of the study cannot be excluded, their impact would have likely been minimal. The candidate SNPs under investigation were all directly typed (i.e. data imputation was not performed). Furthermore, as the rare allele at TNFA −308 is more common among Caucasians than among individuals of Japanese, Chinese, or African ancestry, the accidental inclusion of non-Caucasian cases in our study would have likely biased our OR towards, rather than away, from the null hypothesis or association.
In conclusion, our finding of a significant association between TNFA −308 g>a and primary ITP in Caucasian adults highlights the merits of further research into genetic contributions to the Th1/Th2 imbalance in the disease. This study represents the largest investigation of SNPs in primary ITP to date. Its primary limitation of suboptimal power therefore provides a convincing rationale for the development of an international registry for adults with primary ITP, which could overcome this obstacle. Such a registry would enable testing of reported SNP-disease associations while creating a necessary foundation for the long-term goal of genome-wide association studies to better understand the aetiology of this complex, multi-factorial disease.
The authors thank Dr Charles A Mein for overseeing the genotyping of primary ITP DNA samples, Drs Lynne Condreay and Andres Brainsky for their critical review of the manuscript, and the Wellcome Trust Case−Control Consortium for access to genome-wide data on the 1958 British Birth Cohort. The authors are additionally indebted to our student research assistants: Drs Sarene Saw, Tie Sing Fong, Helen Ngu, Wan Jun Ng, Ei Leen Lim, and Sazlyna Mohd Sazily Lim for their assistance in extracting data from hospital medical records. Lastly, this work would not have been possible without the collaboration of haematologists throughout the United Kingdom. The investigation was supported in part by the UK ITP Support Association, Amgen Europe, and GlaxoSmithKline. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under awards 076113 and 08475.
References
- Cines DB, Blanchette VS. Immune thrombocytopenic purpura. N Engl J Med 2002;346:995–1008.
- Neylon AJ, Saunders PW, Howard MR, Proctor SJ, Taylor PR. Clinically significant newly presenting autoimmune thrombocytopenic purpura in adults: a prospective study of a population-based cohort of 245 patients. Br J Haematol 2003;122:966–74.
- Cohen YC, Djulbegovic B, Shamai-Lubovitz O, Mozes B. The bleeding risk and natural history of idiopathic thrombocytopenic purpura in patients with persistent low platelet counts. Arch Intern Med 2000;160:1630–8.
- Van Eden W, Van Der Zee R, Van Kooten P, Berlo SE, Cobelens PM, Kavelaars A, et al.. Balancing the immune system: Th1 and Th2. Ann Rheum Dis 2002;61(Suppl 2):ii25–8.
- Semple JW. Immune pathophysiology of autoimmune thrombocytopenic purpura. Blood Rev 2002;16:9–12.
- Wang T, Zhao H, Ren H, Guo J, Xu M, Yang R, et al.. Type 1 and type 2 T-cell profiles in idiopathic thrombocytopenic purpura. Haematologica 2005;90:914–23.
- Panitsas FP, Theodoropoulou M, Kouraklis A, Karakantza M, Theodorou GL, Zoumbos NC, et al.. Adult chronic idiopathic thrombocytopenic purpura (ITP) is the manifestation of a type-1 polarized immune response. Blood 2004;103:2645–7.
- Semple JW, Milev Y, Cosgrave D, Mody M, Hornstein A, Blanchette V, et al.. Differences in serum cytokine levels in acute and chronic autoimmune thrombocytopenic purpura: relationship to platelet phenotype and antiplatelet T-cell reactivity. Blood 1996;87:4245–54.
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al.. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 2007;39:1329–37.
- Ates O, Hatemi G, Hamuryudan V, Topal-Sarikaya A. Tumor necrosis factor-alpha and interleukin-10 gene promoter polymorphisms in Turkish rheumatoid arthritis patients. Clin Rheumatol 2008;27:1243–8.
- Waldron-Lynch F, Adams C, Amos C, Zhu DK, McDermott MF, Shanahan F, et al.. Tumour necrosis factor 5' promoter single nucleotide polymorphisms influence susceptibility to rheumatoid arthritis (RA) in immunogenetically defined multiplex RA families. Genes Immun 2001;2:82–7.
- Eliopoulos DG, Mavroudi I, Pontikoglou C, Ximeri M, Stavroulaki E, Pyrovolaki K, et al.. The -509C/T polymorphism of transforming growth factor-beta1 is associated with increased risk for development of chronic idiopathic neutropenia. Eur J Haematol 2009;83:535–40.
- Parks CG, Cooper GS, Dooley MA, Treadwell EL, St Clair EW, Gilkeson GS, et al.. Systemic lupus erythematosus and genetic variation in the interleukin 1 gene cluster: a population based study in the southeastern United States. Ann Rheum Dis 2004;63:91–4.
- Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med 1997;337:1720–5.
- Rodeghiero F, Stasi R, Gernsheimer T, Michel M, Provan D, Arnold DM, et al.. Standardization of terminology, definitions and outcome criteria in immune thrombocytopenic purpura of adults and children: report from an international working group. Blood 2009;113:2386–93.
- Foster CB, Zhu S, Erichsen HC, Lehrnbecher T, Hart ES, Choi E, et al.. Polymorphisms in inflammatory cytokines and Fcgamma receptors in childhood chronic immune thrombocytopenic purpura: a pilot study. Br J Haematol 2001;113:596–9.
- Satoh T, Pandey JP, Okazaki Y, Asahi A, Kawakami Y, Ikeda Y, et al.. Single nucleotide polymorphism of interleukin-1beta associated with Helicobacter pylori infection in immune thrombocytopenic purpura. Tissue Antigens 2009;73:353–7.
- Suzuki T, Matsushima M, Shirakura K, Koike J, Masui A, Takagi A, et al.. Association of inflammatory cytokine gene polymorphisms with platelet recovery in idiopathic thrombocytopenic purpura patients after the eradication of Helicobacter pylori. Digestion 2008;77:73–8.
- Rocha AM, De Souza C, Rocha GA, De Melo FF, Saraiva IS, Clementino NC, et al.. IL1RN VNTR and IL2-330 polymorphic genes are independently associated with chronic immune thrombocytopenia. Br J Haematol 2010;150:679–84.
- Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 2004;96:434–42.
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer 2009;9:361–71.
- Bouma G, Crusius JB, Oudkerk Pool M, Kolkman JJ, von Blomberg BM, Kostense PJ, et al.. Secretion of tumour necrosis factor alpha and lymphotoxin alpha in relation to polymorphisms in the TNF genes and HLA-DR alleles. Relevance for inflammatory bowel disease. Scand J Immunol 1996;43:456–63.
- Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A 1997;94:3195–9.