Abstract
MicroRNAs (miRNAs) are about 19–24 nucleotide small single-stranded noncoding RNAs that are involved in crucial cell processes such as proliferation, apoptosis, and differentiation. Several studies reported show the involvement of miRNA in cancer. It has been suggested that miRNA profiling has the potential to classify a variety of tumors and possibly predict outcome. MicroRNA can act as an oncogene as well as tumor suppressor gene and this dual function of miRNA can be utilized as a therapeutic tool. The oncogenic character of miRNA can be silenced through various RNA interference-type strategies. The involvement of miRNA in the tumorogenesis processes makes them an important therapeutic tool and a novel biomarker. In this review, we have highlighted the role of miRNA in hematological malignancies and its utility in targeted therapy.
Introduction
MicroRNAs (miRNA) are small, endogenous, single-stranded noncoding RNAs, which are about 19–24 nucleotides in length that downregulate gene expression during various crucial cell processes such as proliferation, apoptosis, and differentiation.Citation1 They pair with target transcripts of protein-coding genes and induce RNA cleavage of posttranscriptional inhibition. miRNAs modulate gene expression either by degrading the mRNAs or by blocking translation without degrading the targets.Citation2 Initially, with the discovery of lin-4 that encodes an approximately 22 nucleotide noncoding RNA in Caenorhabditis. elegans, it was observed that they were involved in the larval development of this nematode.Citation3 These noncoding RNAs bind to the partial antisense complementary sequences found in the 3′-untranslated region (3′-UTR) of protein coding gene and repress its translation.Citation1 Several studies have been carried out in plants, fungi, and worms to reveal the role of RNA in mediating silencing of genes homologous to the RNA trigger, a phenomenon generally known as RNA interference. In 1999, Hamilton and BaulcombeCitation4 discovered 22–25 nucleotide RNAs in plants undergoing RNA-mediated gene silencing, which were later named as small interfering RNAs (siRNAs) by Elbashir et al.Citation5 Soon thereafter Mourelatos et al.Citation6 reported the existence of >100 endogenous 22 nucleotide RNAs in different organisms. These RNAs are derived from longer hairpin-like precursors and were named miRNAs.
Each miRNA can control many target genes with different functions, such as transcription factors (TFs), receptors, and transporters.Citation7 With the integral part that miRNA play in cell cycle control, it was logical to suppose that they would play an important role in cancer and leukemia.Citation8 Although the first miRNA was described in 1993, the breadth and diversity of this gene class have been uncovered only in the last few years. At present, several human miRNAs have been reported.Citation9 MicroRNAs are transcribed by the RNA polymerase II enzyme to produce a primary-microRNA (pri-miRNA), which usually contains several kilo bases and is usually capped at the 5′ end and poly-adenylated at the 3′ end, similar to protein-coding mRNAs.Citation10 Several miRNAs have been identified that may regulate the expression of as many as one-third to one half of all protein encoding genes. With the identification of hundreds of miRNAs throughout the genome, the location and function of miRNA could play a significant role in characterization of normal as well as tumor cells.Citation11
MicroRNA processing
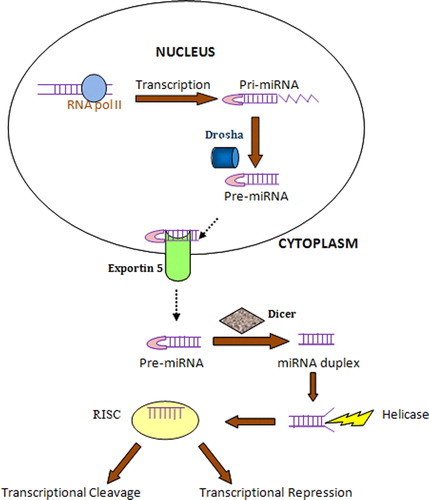
MicroRNAs are generally expressed as polycistronic transcriptsCitation8 and are encoded in intergenic chromosomal regions or can also be found in sense or antisense orientation within the introns of protein coding genes.Citation12 Each miRNA is derived from the stem of a hairpin-like precursor approximately 75 nucleotides (pre-miRNA), which is usually present as a single copy in the genome.Citation3 miRNAs are transcribed as regions of longer RNA molecules that may be as long as 1000 nucleotides. miRNA biogenesis and maturation are basically compartmentalized into nucleus and cytoplasm, respectively.Citation13 The nuclease(s) that processes pri-miRNAs and the factors that export them to the cytoplasm are unknown. miRNAs are first transcribed in the nucleus as primary transcripts (pri-miRNA) some of which are very large, at several kilobases in length with a characteristic stem loop structure and include polycistronic transcripts encoding multiple miRNAs. Analysis of the pri-miRNA precursors have shown that they bear 5′ 7-methyl guanosine cap and a 3′ poly-A tail.Citation14 Pri-miRNA transcripts are transcribed by RNA polymerase II or III using either independent promoters or, as some are found in the introns of protein-encoded genes, they may use the promoter of the proximal coding gene.Citation11
The pri-miRNA is further processed in the nucleus by the Drosha-DGCR8 microprocessor complex to generate a shorter, approximately 70 nucleotide pre-miRNA, which is then transported to the cytoplasm by exportin 5, a member of nuclear transport receptor family.Citation12 Exportin 5 binds to pre-miRNA and the GTP-bound form of the cofactor Ran in the nucleus and releases pre-miRNA following GTP hydrolysis in the cytoplasm.Citation14 In the cytoplasm, DICER, an RNA III-like enzyme that processes dsRNA into siRNAs, excises an imperfectly paired approximately 22 bp double-stranded mature miRNAs from pre-miRNAs. Only one strand of the two strands of a pre-miRNA accumulates in cells as mature miRNA which is one of the key questions in this field to be accomplished. Following Dicer cleavage, the resulting approximately 22 bp double-stranded mature miRNAs is loaded onto an Ago (Agronaute)-2 protein by TRBP (HIV transactivating response RNA-binding protein) to form the RNA-induced silencing complex (RISC). The Ago-2 protein acts as an effector that initially cleaves the miRNA passenger strand and uses the remaining mature single-stranded miRNA as a template of the target mRNA.Citation15 miRNA can direct the RISC to downregulate gene expression by either mRNA cleavage or translational repression.Citation16 ()
Figure 1. MicroRNA biogenesis and processing. The maturation of miRNA from pre-miRNA takes place from nucleus to cytoplasm. The transcription of large pri-miRNA takes place in the nucleus with the help of RNA polymerase II. Pri-miRNA is then cleaved by Drosha, RNase III enzyme to yield pre-miRNA which is then transported to cytoplasm with the help of exportin 5. In the cytoplasm, the hairpin structure is cropped off by the RNase III enzyme, Dicer, producing the double-stranded miRNA duplex which is cleaved by Helicase and incorporated into the RNA-induced silencing complex.
Role of miRNA in normal hematopoiesis
Several studies have shown that miRNAs are involved in various important cell processes ranging from cell proliferation and cell death during development to stress resistance, fat metabolism, insulin secretion, and hematopoiesis. miRNAs play a functional role in both embryonic stem cells tissue somatic stem cells. Hematopoietic system represents a unique biologic tool to evaluate the expression and the functional role of miRNAs in the control of gene expression during normal hematopoietic differentiation.Citation15 TFs are the key regulators of gene expression in multiple cell fate decisions that govern hematopoietic differentiation. TFs and miRNA act together to regulate gene expression during hematopoietic differentiation; TFs regulate the expression of miRNA genes as TFs are the important targets of miRNA.Citation12
The importance of miRNAs in hematopoiesis was revealed by the identification of three murine hematopoietic tissue-specific miRNAs, miR-181a, miR-142s, and miR-223. miR-181a was found to be preferentially expressed in the B-lineage. miR-142 was found to be expressed in B- and myeloid lineages, and miR-223 expression was found to be confined to myeloid lineages.Citation17 Ectopic expression of miR-181a in hematopoietic precursor cells resulted in a doubling of B-lineage cells and paucity of T lymphocytes. However, enforced expression of miR-142 and miR-223 significantly enhance the T-cell count without effecting B-cell numbers.Citation18 In one of the studies, Monticelli et al.Citation19 revealed specific differences between related cell types by pairwise comparisons of the expression of 181 mature miRNAs in selected highly purified hematopoietic cell types at immature, mature, and effector stages. Even though only selected hematopoietic cell lineages were analyzed, each differentiation step was characterized by changes in miRNA expression, with some miRNAs showing increased and some showing decreased expression. It was observed that these miRNAs probably play a role in establishing and/or maintaining cell identity in lymphocytes.
In one of the studies, it has been observed that multiple miRNAs, including miRNA-17, -24, -146, -155, -128, and -181, may hold early hematopoietic cells at an early stem-progenitor stage, blocking their differentiation to more mature cells. miRNA-16, -103, and -107 may block differentiation of later progenitor cells; miRNA-221, -222, and -223 most likely act to control terminal stages of hematopoietic differentiation. miRNA-155 target multiple hematopoietic differentiation-associated molecules, including C/EBPβ, CREBBP, JUN MEIS1, PU.1, AGTR1, AGTR2, and FOS. In addition, it has been reported that miRNA-155 is over expressed in undifferentiated CD34+/CD38− acute myeloid leukemia (AML) stem cells and that ectopic over-expression of miRNA-155 in AML cells blocks differentiation. MicroRNA expression analysis of human CD34+ hematopoietic stem cell precursors from bone marrow and mobilized peripheral blood identified a common signature of 33 miRNAs.Citation20 It has been found that miR-17-5p, -20a, and -106a show an inhibitory effect on monocytic differentiation and maturation by targeting the TF AML1.Citation21 miR-130a targets the TF MAFB that is involved in the activation of GPIIB promoter, a protein for platelet physiology.Citation22 MicroRNA-induced apoptosis of pro-B cell results in enforced expression of miR-150 which, in turn, impairs the lymphocyte development. Downregulation of miR-221 and miR-222 in erythropoietic cells was observed at different stages of erythrocyte differentiation and maturation.Citation23
Importance of miRNA in hematological malignancies
Leukemia is a cancer of the blood or bone marrow characterized by pathological proliferation of abnormal clones of blood cells. It is characterized by either irreparable loss of function of tumor suppressor gene or activation of oncogenes.Citation24 The involvement of miRNA in cancers was first identified by the correlation of genomic localizations of miRNAs and cancer-associated genomic regions (CAGRs).Citation25 Currently, almost all the miRNA-related studies on cancers are based on the different expression profile of miRNAs in cancer cells vs. normal cells. Knockdown or overexpression of a specific miRNA allows studying the specific roles of the miRNA in tumorigenesis. Several groups have studied the expression of miRNA in cancer patients and have observed that miRNAs are expressed differentially in normal and tumor tissues.Citation26 A high proportion of miRNA genes are encoded in the CAGRs, fragile sites, and regions associated with loss of heterozygosity, amplification, and common breakpoint regions. The first proof that miRNAs are involved in cancer came from the finding that miR-15a and miR-16-1 clustered in the 13q14 locus are downregulated or deleted in most patients with chronic lymphocytic leukemia (CLL).Citation27 Both miR-15a and 16-1 target BCL2, antiapoptotic protein which is overexpressed in CLL patients. Transgene expression of miR-15a or miR-16 led to a decrease in the levels of BCL2 protein and the induction of apoptosis in leukaemic cell lines, suggesting that these miRNAs are involved in regulating BCL2 and that their loss in B-CLL may be contributing to the ability of tumor cells to avoid apoptosis.Citation28 The BCR-ABL1 is a hallmark of chronic myelogenous leukemia. In one of the studies, Venturini et al.Citation29 have observed the upregulation of miR 17-92 cluster more in CD34+ cells from chronic phase patients as compared with blast crisis. It has been identified that the expression of miR-17-92 is related to the expression of c-Myc gene. Both miR-17-92 and c-Myc regulate the expression of cell cycle TF gene E2F1. miR-128b, -204, -218, -331, -181b-1 were found to be associated and highly expressed in acute lymphoblastic leukemia patients.Citation30 AML is a most common acute leukemia in adult characterized by rapid proliferation of clonal myeloid precursors that accumulates in the bone marrow and interferes with the normal hematopoiesis.Citation31 MicroRNAs such as miR-15a, -15b, -16-1, let-7a-3, let-7c, let-7d (member of let-7 family), miR-223, -342, and -107 were found to be upregulatedCitation32 ().
Table 1. MicroRNA associated with different hematological malignancies
MicroRNA profiling
Several healthy and pathological tissues have been screened to understand the expression profiling of miRNAs in various diseases. Different expression pattern has been observed in different tissues that encodes the developmental history of the disease. Finding differences in the expression of miRNAs between healthy and diseased cells could potentially be used to diagnose diseases or to assess treatment effects. Number of techniques are available for miRNA profiling, one such technique is oligonucleotide microarray-based analysis that was first started in 2004.Citation38 It is the most common technique that has widely been used to determine the profile of a large number of samples. Bead-based flow cytometry is another technique with high throughput and higher specificity as compared with microarray. It is found to be more accurate and of low cost.Citation39 Deep sequencing is the more recent technique that enables the simultaneous sequencing of different RNA in a single sample. Microarray-based profiling is susceptible to cross hybridization whereas deep sequencing works independently without any prior sequence information.Citation40
MicroRNA as therapeutic tools
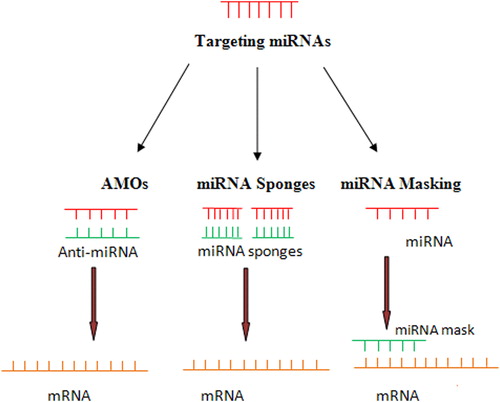
The principle behind the use of miRNA as a therapeutic tool is to restore the expression level of miRNA to normal. At present, thousands of miRNA have been screened for their pathological role in various cancers. Such wide knowledge of miRNA makes them potential tools in therapeutics. miRNA have been found to exhibit dual character, as an oncogene or tumor suppressor gene in several hematological cancers with pathological role in tumor cell proliferation and metastatic process.Citation41 The oncogenic character of miRNA can be silenced through various RNA interference-type strategies, such as the use of anti-miRNA oligonucleotides (AMOs), miRNA sponges, and miRNA masking.
AMOs, also referred to as ‘antagomirs’, are synthetic single-stranded antisense oligonucleotides of 17–22 nucleotides in length that inhibit the interaction of miRNA with its target mRNA. Locked nucleic acid, 2′-O-methoxyethyl AMOs, and 2′-O-methyl AMOs are commonly used AMOs. Despite potential therapeutic agent, AMOs have several drawbacks such as the inability to bind more than one miRNA and their transient duration of action.Citation42
MicroRNA sponges: miRNA sponges were developed to overcome the drawbacks of AMOs and bind the clustered miRNA at more than one point. They were found to interact effectively with the corresponding miRNA and prevent their interaction with the target mRNA. miRNA sponges are effective against all closely related miRNAs within a family with overlapping and redundant targets.Citation43
MicroRNA masking: miRNA masking is another knockdown strategy to the AMOs approach. miRNA mask binds directly to the target miRNA-binding site in the 3′-UTR of its mRNA target.Citation44 Xiao et al.Citation45 utilized the miRNA's principle of actions in a gene-specific manner by an miRNA-masking antisense approach. They designed antisense oligodeoxynucleotides with locked 5′ and 3′ ends, entirely complementary to the miRNA target motifs in the 3′-UTR region. The masking antisense forms duplex with the target mRNA to mask the binding site and stops the action of the miRNA ().
Conclusions
The study of miRNA on gene function in a number of areas has shown them to be a novel technique of gene expression and regulation. Emerging technology and advances in miRNA research have provided detailed knowledge about miRNA biogenesis, mode of action, and the involvement of miRNA in several diseases. miRNA expression through various profiling techniques can successfully identify novel miRNA genes by new technologies thus further enhancing the scope of miRNA functions and therapeutic applications. MicroRNA has been found to be a promising tool for drug development and to be used as a therapeutic tool. The advancement in research of these small nucleotides (miRNA) can be a promising tool in future therapeutic intervention in various cancers.
References
- Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends in molecular medicine. Review. 2006;12(12):580–7.
- Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. Genomics. 2007;8:166.
- Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Biochem. Sci. 2003;28(10):534–40.
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–2.
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200.
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8.
- Albano F, Anelli L, Zagaria A, Liso V, Rocchi M, Specchia G. MIRN 199B downregulation in chronic myeloid leukaemia is associated with deletions on der (9). Br J Haematol. 2008;144:263–75.
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J. Pathol. 2011;223:102–15.
- Griffiths JS, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acid Res. 2008;36:D154–8.
- Dong C, Ji M, Ji C. microRNAs and their potential target genes in leukemia pathogenesis. Cancer Biol. Therapy. 2009;8(3):200–5.
- Huppi K, Volfovsky N, Mackiewicz M, Runfola T, Jones TL, Martin SE, et al. MicroRNAs and genomic instability. Semin Cancer Biol. 2007;17:65–73.
- Navarro F, Lieberman J. Small RNAs guide hematopoietic cell differentiation and function. J Immunol. 2010;184:5939–47.
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell. Biol. 2005;6:376–85.
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–5.
- Pelosi E, Labbaye C, Testa U. MicroRNAs in normal and malignant myelopoiesis. Leuk Res. 2009;33:1584–93.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6.
- Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Manero GG, Calin GA. MicroRNAs and noncoding RNAs in haematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 2008;22:1095–105.
- Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biology. 2005;6(8):R71.
- Georgantas RW, Hildreth R, Morisot S, Alder J, Liu C, Heimfeld S, et al. CD34+ hematopoietic stem progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–5.
- Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–87.
- Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–83.
- Kluiver J, Kroesen BJ, Poppema S, van den Berg A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–6.
- Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–11.
- Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J Clin Invest. 2007;117(8):2059–66.
- Manikandan J, Aarthi JJ, Srinivasan DK, Pushparaj PN. Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2(8):330–4.
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6.
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–9.
- Venturini L, Battmer K, Castoldi M, Schultheis B, Hochhaus A, Muckenthaler MU, et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109:4399–405
- Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva-Jr WA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40(11):1435–40.
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341(14):1051–62.
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–71.
- Marton S, Garcia MR, Robello C, Persson H, Trajtenberg F, Pritsch O, et al. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22:330–8.
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(26):11590–3.
- Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by upregulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902.
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14.
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506.
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101(26):9740–4.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–15.
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
- Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Therapy. 2007;15(12):2070–9.
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20(19):858–61.
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR-430. Science. 2007;318:271–4.
- Xiao J, Yang B, Lin H, Lu Y, Luo X, Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J Cell Physiol. 2007;212(2):285–92.