Abstract
Introduction
The aim of the study was to evaluate the genetic variability of the GP6 gene in patients with sticky platelet syndrome (SPS), a disorder characterized by platelet hyperaggregability, and thus to identify the genetic changes of the glycoprotein VI with possible relation to the platelet hyperaggregability.
Patients and methods
Seventy-one patients with SPS, clinically manifested as ischemic stroke, and 77 controls without SPS and with negative personal history of thromboembolic events were involved. SPS was diagnosed by platelet aggregometry (PACKS-4 aggregometer, Helena Laboratories) according to the method and criteria described by Mammen and Bick. Seven single-nucleotide polymorphisms (SNPs) of the GP6 gene (rs1654410, rs1671153, rs1654419, rs11669150, rs1613662, rs12610286, and rs1654431) were evaluated with the use of restriction-fragment-length polymorphism analysis.
Results
All allele and genotype frequencies were comparable between both SPS patients and the control group with no statistically significant differences. The haplotype analysis showed a higher occurrence of the one major haplotype (TTGTGA, 0.228 vs. 0.174; odds ratio (OR) 1.421; confidence interval (CI) 0.799–2.526) and two minor haplotypes (CGATAA, 0,026 vs. 0,006; OR 4.117; CI 0.443–38.25; TTGTGG, 0.018 vs. 0.009; OR 2.107; CI 0.259–17.12) in patients with SPS. None of haplotype differences was statistically significant. However, both the allele G of SNP rs12610286 (P = 0.029; OR 2.411; CI 1.134–5.123) and one major haplotype (TTGTGA; P = 0.012; OR 2.749; CI 1.223–6.174) were found significantly more frequent in patients with SPS type I in comparison with controls.
Conclusion
Our results, especially higher occurrence of four haplotypes in SPS patients, can support an idea that variability of the GP6 gene may be associated with the platelet hyperaggregability in SPS.
Introduction
Sticky platelet syndrome (SPS) is an inherited thrombophilic disorder, characterized by platelet hyperaggregability after platelet activation with low concentrations of epinephrine (EPI) and/or adenosine diphosphate (ADP).Citation1 According to several studies, SPS is a rather common cause of thrombophilia; some authors claim it to be the second most common cause of hereditary thrombophilia after inherited resistance to activated protein C.Citation2,Citation3 It seems to be particularly frequent among patients with arterial thrombotic events and women with recurrent spontaneous abortions or fetal loss syndrome.Citation4 According to the laboratory findings, three types of the disorder (type I – hyperaggregability after ADP and EPI; type II – hyperaggregability only after EPI; type III – hyperaggregability only after ADP) can be distinguished. Since both genders are affected, SPS has a clear autosomal pattern of inheritance, although the exact genetic cause has not been identified yet. It has been suggested that the defects of the platelet membrane glycoproteins or intracellular signal pathways involved in platelet activation and aggregation are responsible for the disorder.Citation4,Citation5
Ischemic stroke (IS) is one of the commonest clinical manifestations of the syndrome, appearing even during childhood.Citation3,Citation5
Glycoprotein (GP) VI, a member of Ig superfamily, is a 58-kDa platelet transmembrane glycoprotein consisting of 319 amino acids and present on platelet membrane in non-covalent complex with FCRgama subunit.Citation6 It serves, together with integrin complex alpha2beta1, as a receptor for collagen and thus plays an important role in platelet adhesion. The GPVI was shown by several in vitro and in vivo studies to be essential for activation of the integrin for stable adhesion and subsequent signal transduction (via activation of phosphatidylinositol-3-kinase and phospholipase Cgamma2), that leads to granule release, activation of the GPIIb/IIIa via inside-out signaling, and platelet aggregation.Citation7
GPVI is a product of the GP6 gene, localized on chromosome 19 (19q13.4).Citation8 Since the identification and analysis of the GP6 gene in the 1990s numerous single-nucleotide polymorphisms (SNPs) of the gene have been identified.Citation9 Several polymorphisms are known to be associated with increased risk of arterial thrombotic events, namely myocardial infarction and IS.Citation10–Citation13 However, the clinical importance for hemostasis, if any, of the majority of polymorphisms is not yet clear.
The aim of the presented study was to evaluate the genetic variability of the GP6 gene both in the control group and patients with the SPS manifested as IS, carry out association analysis of the GPVI with the syndrome and its types, and thus identify the genetic changes of the GPVI with possible role in platelet hyperaggregability.
Patients and methods
Study population and inclusion/exclusion criteria
The study was approved by the local ethical committee. Informed consent was obtained from each participant or (in case of a person less than 18 years old) from their parents or guardians prior to their involvement in the study.
All patients were initially examined and tested at the hematology laboratory. They were referred to the laboratory by their neurologist or other specialist in order to undergo thrombophilia screening as a part of the differential diagnosis of IS. Patients with verified IS and SPS (according to the criteria mentioned below) were subsequently asked to participate in genotype testing. Thus, all patients fulfilled the following inclusion criteria: (1) at least one episode of IS, as defined in the guidelines of the American Heart Association/American Stroke Association Council on Stroke;Citation14 (2) confirmation of SPS according to the criteria of MammenCitation3 and BickCitation2;, and (3) voluntary signing of informed consent by patient or in case of patient less than 18 years old by their parents or guardians. Exclusion criteria were: (1) unclear or insufficiently verified diagnosis of IS; (2) IS due to the other causes than thromboembolic events or hypercoagulable states; (3) IS of undetermined etiology; and (4) lack of informed consent. For evaluation of the origin of cerebral ischemia the criteria and classification according to Adams et al.Citation15 were used. To exclude a patient, at least one of the exclusion criteria had to be met. Occurrence of other thromboembolic events (e.g. venous thromboembolism, and myocardial infarction) or other thrombophilic states were not considered a reason for patient's exclusion. The patients were examined and recruited in a consecutive manner. Altogether 128 patients were diagnosed with SPS; 71 of them suffered from IS as defined in the inclusion criteria. All 71 suitable subjects agreed to participate in the study. Of those, 15 (21%) had SPS type I, 54 (76%) type II, and 2 (3%) type III ().
Table 1. Characteristics of SPS patients and the control group
Furthermore, 77 randomly chosen healthy individuals were involved as a control group. All controls fulfilled following criteria: (1) negative personal history for IS or other thromboembolic events; (2) negative laboratory testing for SPS; and (3) voluntary signing of informed consent by patient or in case of patient less than 18 years old by his/her tutor ().
Diagnostics of SPS
The blood drawn from antecubital vein, collected into vials prefilled with 3.2% buffered natrium citrate (anticoagulant–blood ratio 1:9), was used for testing of the platelet aggregation. The samples were processed and analyzed within 2 hours after sampling. Platelet aggregability was tested with the use of platelet aggregometry (PACKS-4 aggregometer, Helena Laboratories, Beaumont, TX, USA) according to the method of Mammen.Citation3 Each sample was tested with three low concentrations of ADP (2.34, 1.17, and 0.58 µM) and EPI (11.0, 1.1, and 0.55 µM). The criteria of MammenCitation3 and BickCitation2 were used for the diagnosis and classification of SPS. The testing was performed while the patient did not receive antiplatelet therapy (discontinuation of acetylsalicylic acid or ADP inhibitors for at least 7 days before testing, omitting the use of other drugs with possible effect on platelet activity (e.g. non-steroidal anti-inflammatory drugs) for the same time) and did not suffer from acute thromboembolic events (interval from the last event was 3–5 months).
GP6 gene analysis
For the selection of tag SNPs the data from the International HapMap Project (www.hapmap.org) and an adopted algorithm implemented in Haploview 4.2 (http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview) were used.Citation16 Parameters for Haploview 4.2 were as follows: HapMap Data Release 27 Phase II + III, on NCBI B36 assembly, dbSNP b126, chromosome: 19, region: 60 216–60 242 kb, Analysis Panel: CEU, rCitation2 = 0.8, MAF > 0.1, pairwise tagging. From all 20 SNPs identified with the algorithm, six tag SNPs (rs1654410, rs1671153, rs1654419, rs11669150, rs12610286, and rs1654431) were chosen for analysis. Another, non-tagging SNP (rs1613662) in the coding region of GP6 gene was genotyped as well. The basic characterization of analyzed SNP including nucleotide sequence is given in and their further details are described elsewhere.Citation17,Citation18 The LD plots illustrate structure of relationships between genotyped markers block in CEU HapMap population, SPS, and the control group ().
Figure 1. Relationships between genotyped markers block in CEU HapMap population, SPS patients and the control group (LD plots).
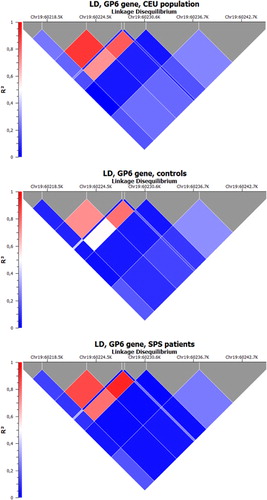
Table 2. Characterization of the examined GP6 SNPs
The blood drawn from antecubital vein, collected into vials prefilled with 3.2% EDTA, was used for DNA analysis. Blood was processed within 2 hours after collection and stored, if necessary, at −20°C. DNA was extracted from peripheral blood leukocytes. Isolation of genomic DNA from the whole blood was performed with SiMaxTM Genomic DNA Extraction kit (SBS Genetech Co., Ltd., Beijing, China) according to the manufacturer's instructions. The GP6 gene polymorphisms were identified with the use of restriction-fragment-length polymorphism and high-resolution melt analysis (in the check on process of one of tag SNPs), with in-house design of individual polymerase chain reaction. Primer3 software was used for the selection of primer sequences (http://frodo.wi.mit.edu/primer3/input.htm). Primers as well as restriction endonucleases used are listed in .
Statistics
Both single marker and haplotype association analysis was performed with HelixTree software (Golden Helix Inc., Bozeman, MT, USA; http://www.goldenhelix.com). Fisher's exact test was employed to estimate significance of deviation from Hardy–Weinberg equilibrium and to execute basic allelic and genotypic association. A P value lower than 0.05 was considered statistically significant. Haplotypes (constructed from six tags SNP) were imputed using EM algorithm, threshold for haplotype frequency estimation was 0.01. Chi-squared test was used for evaluation of haplotype associations. Bonferroni adjustment was used for multiple testing corrections. Overall heterozygosity between groups was evaluated with Kruskal–Wallis test. Throughout the paper, the haplotype is given as a sequence of the alleles of the SNPs in the following order: rs1654410, rs1671153, rs1654419, rs11669150, rs12610286, and rs1654431.
Results
Frequency of all tested alleles and genotypes was comparable between both SPS patients and control group. The frequencies did not differ significantly among groups ().
Table 3. Frequency of alleles of the GP6 SNPs in SPS patients and the control group
The haplotype analysis showed a higher occurrence of one major haplotype (TTGTGA, 0.228 vs. 0.176; odds ratio (OR) 1.421; confidence interval (CI) 0.799–2.526; chi-squared test) and two minor haplotypes (CGATAA, 0.026 vs. 0.006; OR 4.117; CI 0.443–38.25; TTGTGG, 0.018 vs. 0.009; OR 2.107; CI 0.259–17.12; chi-squared test) in patients with SPS in comparison with the control group. However, none of the haplotype differences were statistically significant ().
Table 4. Frequency of haplotypes of the GP6 SNPs (10 most frequent shown) in SPS patients and the control group
The analysis of SPS types found a significantly increased prevalence of the allele G of the SNP rs12610286 in patients with SPS type I, when compared with the control group (P = 0.029; Fisher's exact test; OR 2.411; CI 1.134–5.123) (). The prevalence of all other tested SNPs did not significantly differ among the control group, patients with SPS type I or SPS type II (data not shown). Haplotype analysis showed a significantly increased occurrence of only one major haplotype (TTGTGA; P = 0.012; chi-squared test OR 2.749; CI 1.223–6.174) in patients with SPS type I in comparison with the control group (). Due to the limited number of patients with SPS type III (only 2), that type was not statistically evaluated.
Table 5. Frequency of alleles of the GP6 SNPs in patients with SPS type I and the control group
Table 6. Frequency of haplotypes of the GP6 SNPs (10 most frequent shown) in patients with SPS type I
When overall heterozygosity of selected SNPs in the patient group as well as SPS types was compared with the control group, no association with SPS was detected (P = 0.7076 for the whole patient group; 0.054 for SPS type I; 0.1613 for SPS type II; Kruskal–Wallis test). However, when the heterozygosity of the patients with SPS type I was compared with patients with SPS type II, the difference was significant (P = 0.0045; Kruskal–Wallis test).
Discussion
The GPVI is, according to the present knowledge, a crucial platelet membrane glycoprotein for the adequate platelet activation, adhesion, and aggregation.Citation19 Its role has been studied not only in normal conditions, but in several pathological states characterized by arterial thromboembolism, predominately IS and transient ischemic attack (TIA).Citation20–Citation23 A recent study by Bigalke et al.Citation23 suggests, that GPVI expression on platelet surface is significantly increased in patients with TIA or IS and, furthermore, if severely enhanced, is associated with worse clinical outcome. In last years, with the identification of numerous polymorphisms of the GP6 gene, the question of the impact of genetic changes within the GP6 gene on platelet function emerged. The importance of genetic variability of the GP6 gene for platelet aggregation was stressed by a recent genome-wide meta-analysis by Johnson et al. The analysis focused on the evaluation of the genetic influence on platelet functions and identified seven loci associated with platelet aggregation to physiological agonists (ADP, collagen, and EPI). One of the loci, associated with increased aggregation to collagen, was within the region of the GP6 gene.Citation24 Though a large number of SNPs have been identified in the GP6 gene so far, their exact relation (with few exceptions) to platelet function remains unknown.Citation17,Citation18 This, according to our knowledge, is true for the polymorphisms examined in our study, since their influence on platelet functions in normal or pathological conditions including platelet functional disorders have not been evaluated and published so far.
In our work, we focused on variability of the GP6 gene in patients with inherited thrombophilic platelet disorder, SPS. Our results suggest that global variability of the GPVI gene seems not to be associated with the platelet hyperaggregability in the SPS. Imputed haplotype groups have comparable rates (OR 0.743–1.097) in both SPS patient and control groups, except one major and two minor haplotypes with higher frequencies and one minor haplotype with lower frequencies in SPS patients (OR 0.542), though the differences did not reach the level of statistical significance.
However, when the analysis according to the SPS types was performed, one haplotype was found to be significantly more frequent in patients with SPS type I. This finding suggests that the GP6 SNP rs12610286 in coding region could contribute to the phenotype in SPS in type I, not as a main genetic cause, but as an additional defect.
The combined influence of genetic polymorphisms on platelet function is supported by the observations in several studies, which showed only limited impact of the sole polymorphisms of platelet glycoproteins on platelet function, although overall genetic influence seems to be, as suggested by the testing of siblings and twins, rather high (estimated about 50%).Citation25–Citation27 Recent study by Jones et al.Citation27 analyzing 97 candidate genes involved in collagen and ADP signaling pathways in healthy subjects, showed that interindividual variability was modified by several polymorphisms with, in almost all cases, rather small impact, and not by one sole genetic variation. Analogically, it could be postulated, that the phenotype in some platelet disorders may be modified by complex interaction of several genetic defects and multiple alleles may contribute to the clinical manifestation. Considering phenotype heterogeneity of SPS (hyperaggregability after ADP or EPI, clinical manifestation including arterial and venous thromboembolism), SPS could be, in our opinion, one of these defects.
The limitations of the study – limited number of patients and possible bias in their selection – have to be considered in interpretation of its results. In conclusion, our results, especially higher occurrence of four haplotypes in SPS patients, can support an idea that variability of the GP6 gene may be associated with the platelet hyperaggregability in SPS. Moreover, one of GP6 SNPs, rs12610286, could be involved in platelet hyperaggregability in SPS type I.
Acknowledgements
The study was supported by grants VEGA 1/0018/10, VEGA 1/0029/11 and the European Regional Development Fund (programme ‘Research and Development’, ITMS 262201200016 and 26220120036).
References
- Mammen EF. Ten years' experience with the, sticky platelet syndrome'. Clin Appl Thrombosis/Hemostasis. 1995;1(1):66–72.
- Bick RL. Sticky platelet syndrome: a common cause of unexplained arterial and venous thrombosis. Clin Appl Thromb Hemost. 1998;4(2):77–81.
- Mammen EF. Sticky platelet syndrome. Semin Thromb Hemost. 1999;25(2):361–5.
- Gould WR, Baxi SM, Schroeder R, Peng YW, Leadley RJ, Peterson JT, et al. Gas6 receptors Axl, Sky and Mer enhance platelet activation and regulate thrombotic responses. J Thromb Haemost. 2005;(4):733–41.
- Kubisz P, Ivankova J, Holly P, Stasko J, Musial J. The glycoprotein IIIa PLA1/A2 polymorphism – a defect responsible for sticky platelet syndrome? Clin Appl Thromb Hemost. 2006;12(1):117–9.
- Jandrot-Perrus M, Busfield S, Lagrue AH, Xiong X, Debili N, Chickering T, et al. Cloning, characterization, and functional studies of human and mouse glycoprotein VI: a platelet-specific collagen receptor from the immunoglobulin superfamily. Blood. 2000;96(5):1798–807.
- Clemetson KJ, Clemetson JM. Platelet collagen receptors. Thromb Haemost. 2001;86(1):189–97.
- Clemetson JM, Polgar J, Magnenat E, Wells TN, Clemetson KJ. The platelet collagen receptor glycoprotein VI is a member of the immunoglobulin superfamily closely related to FcalphaR and the natural killer receptors. Biol Chem. 1999;274(41):29019–24.
- Human glycoprotein VI. Available: http://www.genecards.org/cgi-bin/carddisp.pl?gene=GP6&bioalma_dis_art=all&snp=457&rf=/home/genecards/current/website/carddisp.pl#novoseek_dis.
- Cole VJ, Staton JM, Eikelboom JW, Hankey GJ, Yi Q, Shen Y, et al. Collagen platelet receptor polymorphisms integrin alpha2beta1 C807 T and GPVI Q317L and risk of ischemic stroke. J Thromb Haemost. 2003;1(5):963–70.
- Ollikainen E, Mikkelsson J, Perola M, Penttilä A, Karhunen PJ. Platelet membrane collagen receptor glycoprotein VI polymorphism is associated with coronary thrombosis and fatal myocardial infarction in middle-aged men. Atherosclerosis. 2004;176(1):95–9.
- Bray PF, Howard TD, Vittinghoff E, Sane DC, Herrington DM. Effect of genetic variations in platelet glycoproteins Ibalpha and VI on the risk for coronary heart disease events in postmenopausal women taking hormone therapy. Blood. 2007;109(5):1862–9.
- Shaffer JR, Kammerer CM, Dorn J, Ferrell RE, Iacoviello L, Trevisan M, et al. Polymorphisms in the platelet-specific collagen receptor GP6 are associated with risk of nonfatal myocardial infarction in Caucasians. Nutr Metab Cardiovasc Dis. 2011;21(8):546–52.
- Sacco RL, Adams R, Albers G, Alberts MJ, Benavente O, Furie K, et al. Guidelines for prevention of stroke in patients with ischemic stroke or transient ischemic attack: a statement for healthcare professionals from the American Heart Association/American Stroke Association Council on Stroke: co-sponsored by the Council on Cardiovascular Radiology and Intervention: the American Academy of Neurology affirms the value of this guideline. Circulation. 2006;113(10):e409–49.
- Adams HP, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in acute stroke treatment. Stroke. 1993;24(1):35–41.
- Barret JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5.
- Human glycoprotein VI single nucleotide polymorphisms. Available from: http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp&cmd=DetailsSearch&term=GP6+human.
- Human glycoprotein VI single nucleotide polymorphisms. Available from: http://www.ensembl.org/Homo_sapiens/Gene/Summary?g=ENSG00000088053;r=19:55525073-55549632.
- Varga-Szabo D, Pleines I, Nieswandt B. Cell adhesion mechanisms in platelets. Arterioscler Thromb Vasc Biol. 2008;28(3):403–12.
- Penz S, Reininger AJ, Brandl R, Goyal P, Rabie T, Bernlochner I, et al. Human atheromatous plaques stimulate thrombus formation by activating platelet glycoprotein VI. Faseb J. 2005;19(8):898–909.
- Bigalke B, Langer H, Geisler T, Lindemann S, Gawaz M. Platelet glycoprotein VI: a novel marker for acute coronary syndrome. Semin Thromb Hemost. 2007;33(2):179–84.
- Bigalke B, Krämer BF, Seizer P, Fateh-Moghadam S, Gawaz M, Lindemann S. Diagnostic and therapeutic potentials of platelet glycoprotein VI. Semin Thromb Hemost. 2010;36(2):203–11.
- Bigalke B, Stellos K, Geisler T, Kremmer E, Seizer P, May AE, et al. Expression of platelet glycoprotein VI is associated with transient ischemic attack and stroke. Eu J Neurol. 2010;17(1):111–7.
- Johnson AD, Yanek LR, Chen MH, Faraday N, Larson MG, Tofler G, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genetics. 2010;42(7):608–13.
- O'Donnel CJ, Larson MG, Framingham Heart Study. Genetic and environmental contributions to platelet aggregation: the Framingham heart study. Circulation. 2001;103(25):3051–6.
- Hetherington SL, Singh RK, Lodwick D, Thompson JR, Gooddall AH, Samani NJ. Dimorphism in the P2Y1 ADP receptor gene is associated with increased platelet activation response to ADP. Arterioscl Thromb Vasc Biol. 2005;25(1):252–7.
- Jones CI, Bray S, Garner SF, Stephens J, de Bono B, Angenent WG, et al. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009;114(7):1405–16.