Abstract
We previously demonstrated that reactive oxygen species (ROS) could be involved in the DNA damage induced by ultraviolet-C (UVC). In this study, we evaluated singlet oxygen (1O2) involvement in UVC-induced mutagenesis in Escherichia coli cells. First, we found that treatment with sodium azide, an 1O2 chelator, protected cells against UVC-induced lethality. The survival assay showed that the fpg mutant was more resistant to UVC lethality than the wild-type strain. The rifampicin mutagenesis assay showed that UVC mutagenesis was inhibited five times more in cells treated with sodium azide, and stimulated 20% more fpg mutant. These results suggest that 1O2 plays a predominant role in UVC-induced mutagenesis. 1O2 generates a specific mutagenic lesion, 8-oxoG, which is repaired by Fpg protein. This lesion was measured by GC–TA reversion in the CC104 strain, its fpg mutant (BH540), and both CC104 and BH540 transformed with the plasmid pFPG (overexpression of Fpg protein). This assay showed that mutagenesis was induced 2.5-fold in the GC–TA strain and 7-fold in the fpg mutant, while the fpg mutant transformed with pFPG was similar to GC–TA strain. This suggests that UVC can also cause ROS-mediated mutagenesis and that the Fpg protein may be involved in this repair.
Introduction
Sunlight reaching the surface of the Earth possesses a major ultraviolet (UV) component, ultraviolet-A (UVA, 400–320 nm), and a minor component, ultraviolet-B (UVB, 320–290 nm), both of which are known as near-UV radiation. The ultraviolet-C (UVC, 290–100 nm) component, or far-UV, is efficiently filtered by the Earth's ozone layer and atmosphere.Citation1
UV light is directly involved in the induction of basal and squamous carcinomas in humans.Citation2 Many agents, such as UV, which are carcinogens in mammals, also induce lethality of bacterial cells.Citation3 As such, studying the factors involved in bacterial UV damage is of relevance.
Several studies have shown that far-UV (UVC) lethality is generated mainly through the formation of adducts between neighboring pyrimidines (direct lesions), which are lethal if not repaired.Citation4 In contrast, it has been demonstrated that near-UV may generate reactive oxygen species (ROS), which include the hydroxyl radical (·OH), superoxide anion radical (O2·−), hydrogen peroxide (H2O2), and singlet oxygen (Citation1O2).Citation5
Although, hydroxyl radicals cause oxidative modification of DNA bases,Citation6,Citation7 evidence indicates that Citation1O2 is also involved in DNA oxidative damage.Citation8 This ROS is produced by several biological systemsCitation9 and photosensitive reactions, when chromophores are exposed to visible light or are excited by UV light, allowing this energy to be transferred to oxygen and converted into Citation1O2.Citation10
Citation1O2 is known to modify guanine residuals, and is quenched by sodium azide.Citation8 We show that sodium azide protects against the lethality mediated by UVC in Escherichia coli cells, suggesting that DNA damage induced by UVC may involve ROS.
Among the guanine modifications, the main type of DNA lesion induced by Citation1O2 is 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG).Citation11 Although, the lethality of UVC is basically related to direct lesions, other studies have also shown that UVC radiation can induce the formation of oxidative lesions, such as 8-oxoG, in calf thymus DNA.Citation12,Citation13
The Fpg protein (formamidopyrimidine-DNA-N-glycosylase) from E. coli, is a glycosylase, homologous to mammalian OGG1, which excises 8-oxoG linked to cytosine, but not on adenine residue, making up part of the GO (8-oxoG) repair system.Citation14 Citation1O2-induced mutagenicity damage mainly involves base substitutions affecting G:C base pairs. The ability of 8-oxoG to mispair with dG causes GC–TA tranversions.Citation15 Using the CC104 strain created by Cupples and Miller,Citation16 which has a specific mutation in the lacZ gene of E. coli, allowing for the rapid determination of GC–TA tranversion, we saw that UVC induced the 8-oxoG mutagenic lesion mediated by singlet oxygen.
Materials and methods
Bacterial strains
The E. coli strains used in this work are described in .
Table 1. E. coli K-12 bacterial strains and plasmids used in this work
Growth conditions
Cells were grown overnight (in a shaking incubator at 37°C) in LB (Luria-Bertani) medium.Citation17 A starting inoculum was taken from these cultures and cells were grown in the same medium until the mid-exponential growth phase (2 × 108 cells/ml).
Survival experiments
The experiments were performed with cells at the mid-exponential growth phase. The cells grown in LB medium were centrifuged and resuspended in saline (NaCl 0.9%) to the same density, then irradiated with different doses of UVC (254 nm, 15 W general electric G15T8 germicidal lamp, New York, NY, USA). Dosimetry was performed using an ultra-sensitive DC micro-ammeter (Mount Vernon, New York, NY, USA). Samples (0.1 ml) were collected, diluted in saline, and placed (in duplicate) in LB medium solidified with 1.5% agar. The number of colonies formed were counted after incubation overnight at 37°C. Additionally, cells were grown to the mid-exponential phase in LB medium, treated, or not, with sodium azide (100 mM) at the time of UVC irradiation and subsequently removed by centrifugation, and resuspended in saline. The experiments were repeated at least five times and survival data were analyzed using the analysis of variance (ANOVA) test to detect significant differences (P < 0.05) between the tested strains. To identify the differences, the Tukey–Kramer multiple comparison test was employed for all the strains using the InStat statistical program version 4.0 (GraphPad Software, San Diego, CA, USA).
Mutation RifR frequency
In order to determine mutation frequencies, cells at the mid-exponential growth phase were treated, or not, with sodium azide (100 mM) at the time of UVC irradiation and subsequently removed by centrifugation, resuspended in LB medium, and incubated overnight at 37°C. Samples from these cultures were plated on the LB medium containing rifampicin (100 µg/ml). After 2 days incubation at 37°C, the number of rifampicin-resistant mutants (RifR) was scored. The induced mutation frequency was calculated as described by Miller.Citation17 This assay was repeated at least five times and the data were analyzed using ANOVA test, followed by Tukey's multiple comparison test (P < 0.05) using the InStat statistical program, version 4.0.
Alkaline gel eletrophoresis
Alkaline gel electrophoresis was done as described by Zirkle and Krieg,Citation18 with modifications.Citation19 Cells at the exponential growth phase were separated into 1.5 ml samples. The samples were treated, centrifuged (1 minute; 12 000 × g), and suspended in 10 µl cold TE (Tris-EDTA) (50 mM Tris-HCl, pH 8.0; 5 mM ethylenediaminetetra-acetate (EDTA), pH 8.0). Next, 50 µl low-melting-point agarose (1.5%, in TE), at 37°C, was added to each 10 µl sample. The mixture (60 µl) was placed in an acrylic container holding small square wells (0.5 cm2) and incubated for 30 minutes. The blocks were incubated for 2 hours in the dark in a lysing solution (0.1 mM EDTA, pH 8.0; 0.5 M NaOH; 0.05% sodium dodecyl sulfate). They were then washed three times in cold TE (1 ml) and placed onto the electrophoresis gel apparatus with normal agarose solution (0.76%) in an alkaline buffer (0.03 mM NaOH; 0.01 mM EDTA, pH 8.0). After this, the blocks were submitted to 7 V for 14 hours. Then, they were neutralized for 1 hour by soaking in a solution containing 30 mM NaCl and 50 mM Tris-HCl, pH 6.0, stained with ethidium bromide solution (0.5 µg/ml), for 10 minutes, visualized in a UVP transilluminator (Model TM-10E, New York, NY, USA), and digitalized using Canon Power Shot S2IS. The images were analyzed using the Image J software, version 1.33u, in order to quantify the DNA strand breaks. The collected data were submitted to the chi-square test (χCitation2), adopting 5% as the significance level.
E. coli lacZ reversion mutation assay
This mutagenic assay was described by Cupples and MillerCitation16 and very well explained by Josephy.Citation20 Cells were grown overnight in shaking incubators at 37°C in a minimal LB medium with C-saltCitation21 medium enriched with lactose supplemented with 0.5% glucose, 0.2% casaminoacids, and 2 µg/ml thiamine. When the cultures reached the mid-exponential growth phase (2 × 108 cells/ml) they were irradiated with different doses of UVC, centrifuged, and resuspended in the same volume of the C-salt medium without glucose. On the LB plates 0.1 ml aliquots diluted to 10−6 were plated and 0.1 ml on the plates with C-salts enriched with 0.5% lactose and 2 µg/ml thiamine. The plates were incubated at 37°C and screened after 1 day for viable cells, or after 2 days for Lac+ revertants. Only the Lac+ revertants gave readily visible colonies on selective plates. Reversion frequencies were calculated by dividing the number of Lac+ revertants by the total number of viable cells. This assay were repeated at least five times and the data were analyzed using ANOVA test, followed Tukey's multiple comparison test (P < 0.05) using the InStat statistical program, version 4.0.
Results and discussion
UVC radiation is one of the models used to explore the biological consequences of DNA damage, its repair, and tolerance. The damage in question is related to direct damage, such as pyrimidine dimers, which are generated by the interaction of UVC with the cellular genome. However, our results showed that sodium azide conferred protection against the lethal effect induced by UVC in a wild strain of E. coli (, P < 0.001). These data corroborate our previous results, which indicated that ROS mediate UVC-induced lethality in E. coli.Citation22 In this case the protection conferred by sodium azide suggests the involvement of Citation1O2.
Figure 1. Survival of E. coli wild strain (AB1157) (squares) and mutant fpg− (BH20) (triangles) irradiated with different doses of UVC in the presence (filled symbols) or absence (open symbols) of sodium azide. Values are the mean of at least five experiments were submitted to ANOVA test (P < 0.05) followed Tukey's multiple comparison test (P < 0.001).
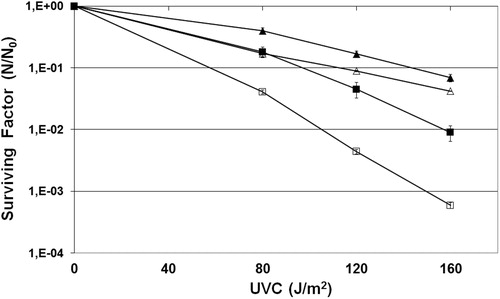
Citation1O2 mainly generates guanine-derived products, like GO lesions, including 8-oxoG and FAPyG.Citation10 The Fpg protein repaired the 8-oxoG in E. coli cells. Thus, a survival assay was done with a mutant strain deficient in Fpg protein (BH20) irradiated with UVC in the presence or absence of sodium azide. The results showed that the BH20 strain was more resistant than the wild-type strain (, P < 0.001). This effect was dose dependent, being more pronounced in higher doses. These results could be explained by the participation of Fpg in the repair mechanism of lesions induced by UVC, which involves the generation of many gaps, besides the ones generated by the nucleotide excision repair (NER) pathway.Citation23 Thus, mutants lacking Fpg repair would produce less DNA breaks than wild-type strains, which would explain the better survival rate of these mutants than wild-type strains.
Many techniques have been developed to detect genotoxicity, such as the comet assay, normally used for eukaryotic cells, and plasmid DNA agarose gel electrophoresis. In order to confirm the participation of 8-oxoG in UVC-induced lethality, alkaline gel electrophoresis was undertaken, which is one of the most widely accepted and used tests in different research areas, including routine genotoxicity assessments and studies of DNA repair processes.Citation24 This single-cell gel electrophoresis technique was developed by Ostling and JohansonCitation25 and Singh et al.Citation26 It is a simple method for measuring DNA strand breaks in eukaryotic cells.
Alkaline gel electrophoresis was performed with a wild strain and the BH20 strain irradiated with UVC in the presence or absence of sodium azide (100 mM) that in the survival control test demonstrated was not lethal to the E. cells (data not shown). First, the gel densitometry analysis revealed that the mutant fpg− displayed fewer strand breaks than the wild strain (P < 0.001), corroborating our previous resultsCitation22 (). This suggested that the wild strain repaired this kind of UVC-induced lesion (8-oxoG), recognizing and removing it by the Fpg protein. This repair process generates a pyrimidinic sitesCitation3 which can result in DNA strand breaks, as shown in the present proposed methodology, which is relatively new. This was not the case for the fpg− mutant.
Figure 2. Alkaline gel electrophoresis of E. coli irradiated with UVC (120 J/m2) in the presence or absence of sodium azide with percentual of DNA stand breaks. Lanes: (1) AB1157; (2) AB1157 + UVC; (3) AB1157 + azide; (4) AB1157 + UVC + azide; (5) BH20; (6) BH20 + UVC; (7) BH20 + azide; (8) BH20 + UVC + azide. The image represent a single experiment and the collected data of three experiments were submitted to the χ2, adopting 5% as the significance level (P < 0.001).
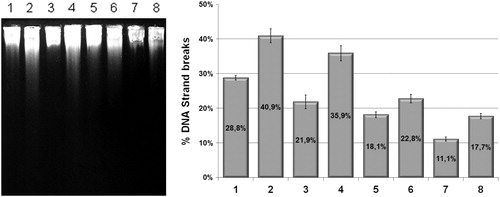
When the cells were treated with sodium azide, the number of strand breaks visible in the gel for both the wild and mutant strains was lower (P < 0.001), confirming that the 8-oxoG could be a lesion induced by UVC and that when sodium azide quenches the Citation1O2 it inhibits the formation of this kind of lesion in DNA (). However, the controls (cells with only sodium azide) also possess less DNA strand breaks, because of its antioxidant potential on quenching spontaneous ROS.
In order to check whether sodium azide diminishes the mutagenic effect induced by UVC, we conducted mutagenesis experiments with wild strain and fpg mutant strain cells irradiated with UVC in the presence or absence of sodium azide. The analyses of rifampicin mutagenesis detected rpoB gene alterations that conferred the bacteria rifampicin resistance. Our results show that sodium azide conferred, in a dose-dependent manner, up to 6-fold protection for the cells against UVC-induced mutagenesis (, P < 0.001). The reduce UVC-induced mutagenesis by sodium azide in the wild strain (, P < 0.001) was due to a reduction in the amount of 8-oxoG generated. This could allow an easier repair, without leading to cell lethality through the DNA repair accumulation.
Figure 3. Mutagenesis RifR of E. coli wild strain (AB1157) (black column) and mutant fpg− (BH20) (gray column) irradiated with different doses of UVC in the presence (white mixed column), or absence of sodium azide. Values are the mean of at least five experiments (P < 0.001).
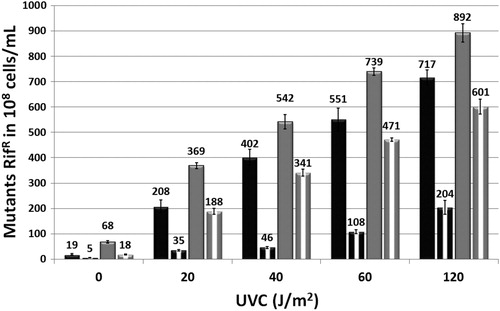
As such, the Citation1O2 produced by the UVC could generate 8-oxoG in the bacterial cells. This could explain the protection obtained against mutagenic effects when the cells were irradiated in the presence of sodium azide. Also, the UVC induced up to 50% more mutagenesis in the fpg mutant than in the wild strain (P < 0.001), corroborating with hypothesis that Fpg participates in the repair of UVC-induced lesions ().
The lacZ reversion mutation assay was performed to verify whether 8-oxoG was involved in the UVC-induced mutagenesis. This method uses mutant strains that have a lacZ mutation resulting from a base substitution at coding position 461 in the lacZ gene. The altered sequences result in a loss of ability to metabolize lactose (Lac− phenotype). The reversion to Lac+ occurs by base substitution, reverting to the proper reading frame for beta-galactosidase synthesis.Citation27,Citation28 The CC104 strain can detect the GC → TA transversion, and this mutagenesis could be induced by 8-oxoG.Citation29 The survival of LacZ strains was statistically the same as the homologous strains used (P > 0.05) (data not shown).
Our results showed that the UVC-induced mutagenesis in the fpg mutant strain BH540 was seven times greater than in the CC104 strain (, P < 0.001). Also, the mutant fpg transformed with plasmid pFpg, returning to its original mutagenesis (P > 0.05). The survival assay of BH20 strain transformed with plasmid pFPG has also not shown difference in UVC sensitivity compared to wild strain (P > 0.05) (data not shown). These results suggest that the fpg gene product (Fpg protein or endonuclease IV) seems to be very important in the repair of UVC-induced DNA lesions, indicating that Citation1O2 mediates the mutagenesis induced by UVC, and that this lesion is probably 8-oxoG, a mutagenic lesion.
Figure 4. Mutagenesis LacZ of E. coli CC104, BH540, CC104 + pFPG, and BH540 + pFPG irradiated with 120 J/m2 UVC (gray column) or not irradiated (gray column). Values are the mean of at least five experiments and standard deviations were calculated (P < 0.001).
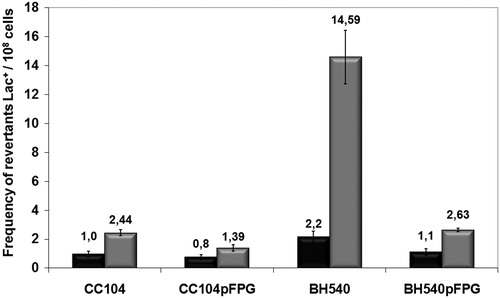
This could indicate the participation of the ROS, Citation1O2, in UVC-induced mutagenic lesions, forming 8-oxoG, a lesion that is repaired by the Fpg protein, a base excision repair (BER) protein. UVC-induced lesions may be repaired using the BER and NER pathway, and these lesions could be repaired simultaneously with other UVC-induced DNA lesions, like pyrimidine dimers.
Acknowledgements
Research supported by CAPES, FAPERJ, CNPq, and UERJ.
References
- Serafini DM, Schellhorn HE. Endonuclease III and endonuclease IV protect Escherichia coli from the lethal and mutagenic effects of near-UV radiation. Can J Microbiol 1999;45:632–7.
- Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst 1981;66:1191–308.
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2nd ed. Washington, DC, USA: ASM Press; 2006.
- Hart RW, Setlow RB, Woodhead AD. Evidence that pyrimidine dimers in DNA can give rise to tumors. Proc Natl Acad Sci USA 1977;74:5574–8.
- Rosen JE, Prahalad AK, Williams GM. 8-oxodeoxyguanosine formation in the DNA of cultured cells after exposure to H2O2 alone or with UVB or UVA irradiation. Photochem Photobiol 1996;64:117–22.
- Cadet J, Teoule R. Comparative study of oxidation of nucleic acid components by hydroxyl radicals, singlet oxygen and superoxide anion radicals. Photochem Photobiol 1978;28:661–7.
- Ames BA, Gold LS. Endogenous mutagens and the cause of aging and cancer. Mutat Res 1991;250:3–16.
- Piette J. Biological consequences associated with DNA oxidation mediated by singlet oxygen. J Photochem Photobiol 1991;11:241–60.
- Briviba K, Klotz LO, Sies H. Toxic and signaling effects of photochemically or chemically generated singlet oxygen in biological systems. Biol Chem 1997;378:1259–65.
- Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. New York, USA: Oxford University Press; 1998.
- Cadet J, Berger M, Douki T, Morin B, Raoul S, Ravanat JL, et al. Effects of UV and visible radiation on DNA – final base damage. Biol Chem 1997;378:1275–86.
- Wei H, Cai Q, Rahn R, Zhang X. Singlet oxygen involvement in ultraviolet (254 nm) radiation induced formation of 8-hydroxy-deoxyguanosine in DNA. Free Rad Biol Med 1997;23:148–54.
- Zhang X, Rosenstein BS, Wang Y, Lebwohl M, Wei H. Identification of possible reactive oxygen species involved in ultraviolet radiation-induced oxidative DNA damage. Free Rad Biol Med 1997;23:980–5.
- Michaels ML, Tchou J, Grollmanj VP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 1997;31:10964–8.
- Menck CFM, Di Mascio P, Agnez LF, Ribeiro DT, Costa de Oliveira R. Genetic deleterious effects of singlet oxygen. Quimica Nova 1993;16:328–36.
- Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA 1989;86:5345–9.
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1992.
- Zirkle RE, Krieg NR. Development of a method based on alkaline gel electrophoresis for estimation of oxidative damage to DNA in Escherichia coli. J Appl Bacteriol 1996;81:133–8.
- Mattos JCP, Motta ES, Oliveira MBN, Dantas FJS, Araujo AC. Alkaline gel electrophoresis assay to detect DNA strand breaks and repair mechanisms in Escherichia coli. Braz Arch of Biol Tech 2008;51:121–6.
- Josephy PD. The Escherichia coli lacZ reversion mutagenicity assay. Mutat Res 2000;455:71–80.
- Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 1956;218(1):97–106.
- Gomes AA, Silva-Júnior ACT, Oliveira EB, Asad LMBO, Reis NCSC, Felzenszwalb I, et al. Reactive oxygen species mediate lethality induced by far-UV in Escherichia coli cells. Redox Rep 2005;10:91–5.
- Caldeira de Araújo A, Favre A. Near ultraviolet DNA damage induces the SOS response in Escherichia coli. EMBO J 1986;5:175–9.
- Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, et al. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen 2000;35:206–21.
- Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damage in individual mammalian cells. Biochem Biophys Res Commun 1984;23:291–8.
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantification of low levels of DNA damage in individual cells. Exp Cell Res 1988;175:184–91.
- Cupples CG, Cabrera M, Cruz C, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 1990;125:275–80.
- Borden A, O'Grady PI, Vandewiele D, Fernandez de Henestrosa AR, Lawrence CW, Woodgate R. Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn Cyclobutane dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J Bacteriol 2002;184:2674–81.
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-hydroxyguanine, an abundant form of oxidative DNA damage, causes G—T and A—C substitutions. J Biol Chem 1992;267:166–72.