Abstract
Reactive oxygen species (ROS) mediate various cell signaling processes, but the mechanism for how ROS promote cell signaling is poorly understood. Protein carbonylation occurs because of the direct metal-catalyzed oxidation of amino acid side chains (primary protein carbonylation) or the addition of reactive aldehydes to amino acid side chains (secondary protein carbonylation). We hypothesize that primary protein carbonylation plays a role in the mechanism of ROS signaling. Specifically, we propose that (i) primary protein carbonylation mediates cell signaling and (ii) primary protein carbonylation is reversible.
Introduction
Reactive oxygen species (ROS) mediate various cell signaling processes,Citation1 but the mechanism for how ROS promote cell signaling is poorly understood. Protein carbonylation occurs because of the direct metal-catalyzed oxidation of amino acid side chains (primary protein carbonylation) or the addition of reactive aldehydes to amino acid side chains (secondary protein carbonylation). We hypothesize that primary protein carbonylation plays a role in the mechanism of ROS signaling. Specifically, we propose that (i) primary protein carbonylation mediates cell signaling and (ii) primary protein carbonylation is reversible.
Protein carbonylation
Protein carbonylation is a term used to describe a process, in which reactive aldehydes or ketones are introduced into proteins by oxidation.Citation2 Primary protein carbonylation involves the direct metal-catalyzed oxidation of amino acid side chains, particularly on four susceptible amino acid residues: proline, arginine, lysine, and threonine.Citation3,Citation4 The oxidation of the proline or arginine side chains results in the formation of glutamic semialdehyde, while lysine oxidation produces aminoadipic semialdehyde, thereby introducing carbonyl groups in the protein structure.Citation5 Oxidation also converts the hydroxyl group of threonine side chains to carbonyl, forming 2-amino-3-ketobutyric acid. Secondary protein carbonylation mechanisms include the Michael addition reactions of reactive aldehydes with lysine, histidine, and cysteine residue side chains, as well as the formation of Schiff bases by reducing sugars with amino groups on lysine and arginine, resulting in an increase in advanced glycation end-products.Citation6,Citation7 Protein carbonyls occur in various oxidative stress and disease conditions.Citation7,Citation8 In addition to proteins being inactivated, carbonylated cellular proteins undergo proteasome-dependent degradation. Primary protein carbonylation is thought to play a role in labeling damaged proteins during oxidative stress in order to eliminate them from the biological system.Citation8
Hypothesis #1: primary protein carbonylation mediates cell signaling
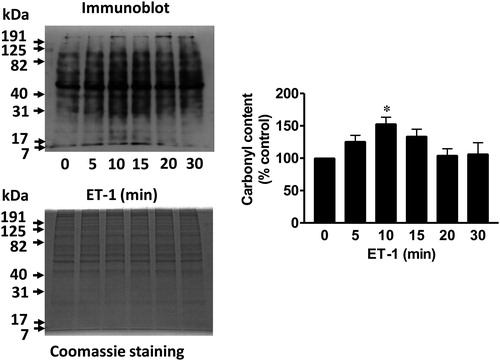
We reported that ligand–receptor stimulation in response to the treatment of vascular smooth muscle cells with endothelin-1 (ET-1) promoted the carbonylation of various proteins as monitored using 2,4-dinitrophenylhydrazine (DNPH) ().Citation9 Mass spectrometry identified some of these proteins, which include annexin A1 and peroxiredoxin.Citation9 Annexin A1 is a protein that functions as an anti-inflammatory, anti-proliferative, and pro-apoptotic factor. We found that subsequent to being carbonylated, this protein is degraded by proteasomes.Citation9 The degradation of annexin A1 should thus promote pro-inflammatory, proliferative, and anti-apoptotic signaling. Thus, we propose that oxidation-dependent proteasomal degradation of annexin A1 drives signaling for cell growth. This mechanism has been to date observed in smooth muscle cellsCitation9 and in heart tissue.Citation10 We recently suggested that this oxidant-mediated process defines the differences in cell signaling mechanisms observed between right and left heart ventricles.Citation10 Specifically, in the right ventricle, pressure overload triggers carbonylation and degradation of annexin A1, which in turn activates the CBF/NF-Y for gene transcription of GATA4, a major regulator of cardiac hypertrophy.Citation10
Figure 1. ET-1 promotes protein carbonylation in smooth muscle cells. Pulmonary artery smooth muscle cells were treated with ET-1 (30 nM) and harvested at the indicated time points. Cell lysates were prepared and derivatized with DNPH. Carbonylated protein levels were monitored by Western blotting with the DNP antibody. Coomassie blue staining showed no differences in total protein levels. The bar graph shows the mean ± SEM of the percentage of total carbonyl content relative to the untreated control as determined by densitometry. * denotes the value that is significantly different from the untreated control at P < 0.05 (n = 6), as determined by one-way ANOVA with a Student–Newman–Keuls post-hoc test. Reproduced from Wong et al. (2008)9 in accordance with the Copyright Transfer Agreement.
In addition to this mechanism, which involves carbonylation-mediated protein degradation, we propose that primary protein carbonylation may also inhibit or activate the activities of various signaling molecules. For example, ET-1-mediated peroxiredoxin carbonylationCitation9 may inhibit the activity of this antioxidant enzyme and, in turn, amplify the production of ROS that are generated by NAD(P)H oxidase. Rhee and co-workersCitation11 recently reported that the inactivation of peroxiredoxin allows localized H2O2 accumulation for cell signaling.
DNPH can label primary as well as secondary carbonyls.Citation12 In addition, experiments by Dalle-Donne et al.,Citation13 using a short peptide containing six amino acids, suggested that DNPH could also react with sulfenic acid. Whether the data of a number of published studies that used DNPH to assess the occurrence of protein carbonylation are confounded by the possible detection of sulfenic acid in protein structures remains unknown. Nevertheless, our data using DNPH may not necessarily be due to the involvement of primary protein carbonylation. The following experimental results should strengthen the hypothesis that observed changes in DNPH adducts during cell signaling events are largely because of primary carbonyls.
To assess the involvement of secondary protein carbonylation, we tested whether ET-1 promotes the formation of protein adducts with 4-hydroxynonenal (4-HNE) and/or malondialdehyde (MDA), major lipid peroxidation products that can produce secondary protein carbonylation. As shown in A, no apparent changes occurred in 4-HNE or MDA protein adduct levels by treating cells with ET-1, during which time a significant modulation of DNPH adducts was detected. Thus, secondary protein carbonylation, at least via these two lipid-derived aldehydes, is not largely responsible for the DNPH adduct formation in response to ET-1.
Figure 2. Evidence to support the hypothesis for the occurrence of primary protein carbonylation in ET-1 signaling. Pulmonary artery smooth muscle cells were treated with ET-1 (30 nM) and harvested at the indicated time points to prepare cell lysates as previously described [Wong et al., 2008]. (A) Cell lysates were subjected to Western blotting using 4-HNE and MDA antibodies. The bar graph shows the means ± SEM (n = 4) of the percentage of total 4-HNE or MDA adduct content relative to the untreated control as determined by densitometry. (B) Cell lysates (20 µg protein) were treated with 1% SDS, derivatized with biotin hydrazide (5 mM) for 2 hours by mixing at room temperature, incubated with sodium cyanoborohydride (15 mM) on ice for 30 minutes and precipitated with trichloroacetic acid (10%) on ice. Protein pellets were washed three times with ethanol:ethyl acetate before loading onto gels for Western blotting using avidin-horseradish peroxidase. * denotes significantly different values from untreated control at P < 0.05 (n = 4), as determined by one-way ANOVA with a Student–Newman–Keuls post-hoc test.
![Figure 2. Evidence to support the hypothesis for the occurrence of primary protein carbonylation in ET-1 signaling. Pulmonary artery smooth muscle cells were treated with ET-1 (30 nM) and harvested at the indicated time points to prepare cell lysates as previously described [Wong et al., 2008]. (A) Cell lysates were subjected to Western blotting using 4-HNE and MDA antibodies. The bar graph shows the means ± SEM (n = 4) of the percentage of total 4-HNE or MDA adduct content relative to the untreated control as determined by densitometry. (B) Cell lysates (20 µg protein) were treated with 1% SDS, derivatized with biotin hydrazide (5 mM) for 2 hours by mixing at room temperature, incubated with sodium cyanoborohydride (15 mM) on ice for 30 minutes and precipitated with trichloroacetic acid (10%) on ice. Protein pellets were washed three times with ethanol:ethyl acetate before loading onto gels for Western blotting using avidin-horseradish peroxidase. * denotes significantly different values from untreated control at P < 0.05 (n = 4), as determined by one-way ANOVA with a Student–Newman–Keuls post-hoc test.](/cms/asset/dc88c937-1d1a-4685-84f6-2bc5f58d7318/yrer_a_11653440_f0002_b.jpg)
Some of our experiments do not support the involvement of sulfenic acid in DNPH adduct formation during cell signaling. Key DNPH signals we selected for the further studies of cell signaling were iron-dependent and inhibited by deferoxamine.Citation9 Since H2O2 has been reported to drive DNPH–sulfenic acid interactions in the presence of a metal chelator,Citation13 it is unclear if such interactions occur in the mechanism that can be inhibited by deferoxamine. We did detect some ET-1-promoted DNPH signals, which did not respond to deferoxamine. These may be sulfenic acid signals detected by DNPH. If so, this also argues that deferoxamine inhibits the downstream components of H2O2, such as the Fenton reaction rather than NAD(P)H oxidase, for suppressing the carbonylation of selected signaling molecules.
Furthermore, according to Dalle-Donne et al.,Citation13 sulfenic acid–DNPH interactions are driven by acidic pH, which is required for DNPH derivatization. We tested whether changes in possible carbonyl signals may be visible using biotin hydrazide as a probe, which does not require acidic conditions to react with carbonyl groups.Citation14 We found that an ET-1-mediated transient increase in the ‘protein carbonylation’ signal was also observed by derivatizing with biotin hydrazide (B).
We thus believe that the observed changes in DNPH adduct levels in response to the ET-1 treatment of cells are because of the changes in the state of primary protein carbonylation, and hypothesize that primary protein carbonylation plays a role in ROS signaling.
Hypothesis #2: primary protein carbonylation is reversible
As described above, we found that ET-1 promotes protein carbonylation. Typically, in pulmonary artery smooth muscle cells, ET-1-mediated carbonylation occurs as early as 5 minutes and peaks at about 10 minutes post-ET-1 stimulation.Citation9 Interestingly, as shown in , this is followed by a decrease in carbonylated proteins within 30 minutes, revealing that the process is transient.Citation9 We named this process ‘protein decarbonylation’. While such transient kinetics are typical for signal transduction processes such as protein phosphorylation/dephosphorylation, it was surprising that primary protein carbonylation, a type of protein oxidation that is usually considered irreversible,Citation3,Citation4 exhibited such kinetics. We initially hypothesized that carbonylated proteins may be degraded, but a decrease in carbonyl content did not coincide with decreased total protein expression () and proteasome inhibition did not suppress this decrease.Citation9 We subsequently found that thiol reductants can decrease the carbonyl content,Citation9 opening up the possibility that primary protein carbonylation may be reversible. We provided evidence for the role of thioredoxin in the mechanism of protein decarbonylation.Citation9
While further work is needed to substantiate this idea and to find the mechanism of reversibility, some theoretical mechanisms can be postulated to explain how primary protein carbonylation can be reversed. For free amino acids, enzymatic pathways reduce carbonylated amino acid products back to their native amino acids. For example, 2-amino-3-ketobutyric acid (l-2-amino-3-oxobutanoic acid) can be reduced to l-threonine by using NADH as an electron donor, which is catalyzed by an enzyme, threonine 3-dehydrogenase, in its reverse reaction.Citation15,Citation16 In addition, aminoadipic semialdehyde can be converted back to lysine via saccharopine with the catalysis of NADH- or NADPH-dependent saccharopine dehydrogenase with glutamate as a co-factor.Citation17,Citation18 Further, glutamic semialdehyde can be reduced to pyrroline-5-carboxylate, which in turn can be reduced to proline by pyrroline-5-carboxylate reductaseCitation19 or proline dehydrogenase.Citation20 In this case, however, its role in protein amino acid reduction may not apply because pyrroline-5-carboxylate is not an amino acid. Via ornithine aminotransferase with glutamate as a co-factor, glutamic semialdehyde can also become ornithine, which can then become arginine.Citation21
Furthermore, at least three classes of enzymes are capable of reducing reactive aldehydes, namely alcohol dehydrogenases, short chain dehydrogenase reductases including carbonyl reductases, and aldo-keto reductases.Citation6 Like the enzymes described above, these enzymes are known to react with small molecule aldehydes, hence it is unclear whether protein amino acids can be substrates for these enzymes. However, possible reactions with protein carbonyls have not yet been excluded.
Conclusion
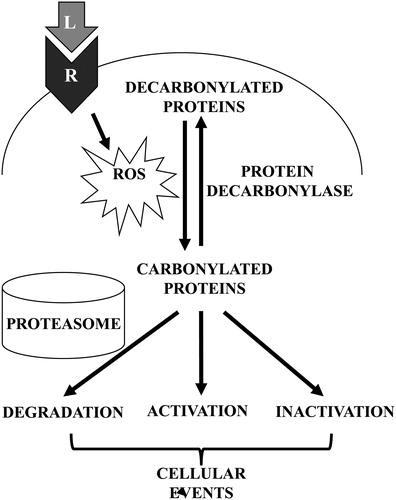
Here, we postulate that primary protein carbonylation may regulate cell signaling events. The consequences of carbonylation may include the direct activation or inhibition of the activities of target proteins as well as selective degradation by proteasomes. The possibility for enzyme-catalyzed, reversible primary protein carbonylation further suggests the potential regulatory role of this process in cell signaling mechanisms. These proposed mechanisms are summarized in .
Figure 3. Schematics of proposed mechanisms involving primary protein carbonylation in cell signaling. Receptor (R) interactions with ligands (L) such as ET-1 trigger the generation of ROS, which in turn promote the metal-catalyzed formation of primary protein carbonyls. Carbonylated proteins may be selectively degraded or may influence the activities of signal transduction molecules to elicit cellular events. Primary protein carbonylation may be reversible by as-yet unidentified protein decarbonylases.
Acknowledgements
This work was supported in part by grants from National Institutes of Health (R01HL72844 and R01HL97514) to Y.J.S.
References
- Suzuki YJ, Forman HJ, Sevanian A. Oxidants as stimulators of signal transduction. Free Radic Biol Med 1997;22:269–85.
- Suzuki YJ, Carini M, Butterfield DA. Protein carbonylation. Antioxid Redox Signal 2010;12:323–5.
- Nyström T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J 2005;24:1311–7.
- Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein carbonylation in human diseases. Trends Mol Med 2003;9:169–76.
- Requena JR, Chao CC, Levine RL, Stadtman ER. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci USA 2001;98:69–74.
- Ellis EM. Reactive carbonyls and oxidative stress: potential for therapeutic intervention. Pharmacol Ther 2007;115:13–24.
- Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res 2010;9:3766–80.
- Levine RL. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic Biol Med 2002;32:790–6.
- Wong CM, Cheema AK, Zhang L, Suzuki YJ. Protein carbonylation as a novel mechanism in redox signaling. Circ Res 2008;102:310–8.
- Park AM, Wong CM, Jelinkova L, Liu L, Nagase H, Suzuki YJ. Pulmonary hypertension-induced GATA4 activation in the right ventricle. Hypertension 2010;56:1145–51.
- Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 2010;140:517–28.
- Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol 1994;233:346–57.
- Dalle-Donne I, Carini M, Orioli M, Vistoli G, Regazzoni L, Colombo G, et al. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radic Biol Med 2009;46:1411–9.
- Yoo BS, Regnier FE. Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis 2004;25:1334–41.
- Aoyama Y, Motokawa Y. L-Threonine dehydrogenase of chicken liver. Purification, characterization, and physiological significance. J Biol Chem 1981;256:12367–73.
- Marcus JP, Dekker EE. Threonine formation via the coupled activity of 2-amino-3-ketobutyrate coenzyme A lyase and threonine dehydrogenase. J Bacteriol 1993;175:6505–11.
- Jones EE, Broquist HP. Saccharopine, an intermediate of the aminoadipic acid pathway of lysine biosynthesis. 3. Aminoadipic semialdehyde-glutamate reductase. J Biol Chem 1966;241:3430–4.
- Saunders PP, Broquist HP. Saccharopine, an intermediate of the aminoadipic acid pathway of lysine biosynthesis. IV. Saccharopine dehydrogenase. J Biol Chem 1966;241:3435–40.
- Smith ME, Greenberg DM. Characterization of an enzyme reducing pyrroline-5-carboxylate to proline. Nature 1956;177:1130.
- Scarpulla RC, Soffer RL. Membrane-bound proline dehydrogenase from Escherichia coli Solubilization, purification, and characterization. J Biol Chem 1978;253:5997–6001.
- Seiler N. Ornithine aminotransferase, a potential target for the treatment of hyperammonemias. Curr Drug Targets 2000;1:119–53.