Abstract
Nine cases of cryptosporidiosis co-infections in AIDS patients were clinically categorised into severe (patients 1, 3, 8 and 9), moderate (patients 4 and 5) and mild (patients 2, 6 and 7). Formalin-fixed faecal specimens from these patients were treated to obtain high quality DNA competent for amplification and sequencing of the 60-kDa glycoprotein (GP60) gene. Sequence analysis revealed that one patient was infected with Cryptosporidium hominis whereas the remaining eight patients were infected with C. parvum. Interestingly, the patients showing severe cryptosporidiosis harboured two subtypes within the C. parvum allelic family IIc (IIcA5G3 and IIcA5G3R2), whereas patients with moderate or mild infections showed various subtypes of the C. parvum allelic family IIa (IIaA14G2R1, IIaA15G2R1, IIaA17G3R1 and IIaA18G3R1).
DNA extraction and genotyping of Cryptosporidium spp. is a challenging task on formalin-fixed stool samples, whose diagnostic outcome is age-dependent. The method herein reported represents a step forward routine diagnosis and improves epidemiology of HIV-related clinical cases. Due to the need to elucidate genetic richness of Cryptosporidium human isolates, this approach represents a useful tool to correlate individual differences in symptoms to subgenotyping lineages.
INTRODUCTION
Cryptosporidium spp. are oocysts-forming Apicomplexan protozoa that cause a widespread enteric infection (cryptosporidiosis) in humans and animals, a disease which results in sickness and severe diarrhoea. In the immunocompetent host, the symptoms, when present, last for about 2 weeks before the infection is cleared (Riggs, Citation2002). Conversely, in high-risk host groups, particularly neonates (Huang et al., Citation2004; Keating, Citation2005) or people with suppressed or deficient immune systems (i.e. HIV/AIDS patients), the infection can be severe, long-lasting and even fatal (Amadi et al., Citation2002; Certad et al., Citation2005). The vast majority of human cases of cryptosporidiosis worldwide are caused by two species, Cryptosporidium parvum and C. hominis (Morgan et al., Citation1999). However, other species, including C. felis, C. meleagridis, C. canis, C. suis, C. muris (Xiao and Ryan, Citation2004) and, more rarely, C. cuniculus (Robinson et al., Citation2010) and C. ubiquitum (Fayer et al., Citation2010) can infect humans too, especially children under the age of 5 years and immunocompromised individuals (Abubakar et al., Citation2007).
The identification and characterization of Cryptosporidium species and population variants (genotypes and subgenotypes) is fundamental to study the epidemiology of cryptosporidiosis, being a valid support for prevention and control strategies (Putignani and Menichella, Citation2010). Oocyst morphology, host specificity or preferences in infection sites do not provide sufficient information for the identification of Cryptosporidium species, genotypes or subgenotypes (Fall et al., Citation2003; Jex and Gasser, Citation2010). Advances in molecular technologies have led to significant improvements in the characterization of the genetic variability among and within Cryptosporidium species (Jex and Gasser, Citation2008). In particular, the amplification and sequencing of one or more genetic loci (markers) have been used for the categorization of Cryptosporidium species, genotypes or subgenotypes (Xiao et al., Citation2004; Chalmers et al., Citation2005; Plutzer and Karanis, Citation2009; Bouzid et al., Citation2010). Mini- and micro-satellites, or simple sequence repeats, constitute a rich source of polymorphism and have been extensively used for high-resolution genotyping and mapping (Feng et al., Citation2000). In particular, the GP60 gene is useful for such studies as it contains multiple regions with high mutation rates, including a ‘hyper-variable’ microsatellite region (Strong et al., Citation2000).
Herein we describe a successful retrospective subgenotyping of C. parvum and C. hominis isolates, performed on aged formalin-fixed stool samples from nine HIV-infected patients, based on a combination of optimized DNA extraction and sequence analysis of a fragment of the GP60 gene. Subgenotype lineages were correlated to clinical manifestations for each patient. The understanding of this relationship may represent the starting point for further extended studies on the correlation between isolate type and symptom severity in HIV-infected patients.
MATERIALS AND METHODS
Collection of Faecal Specimens and Parasite Identification
Formalin-fixed stool samples were collected from nine HIV-infected patients admitted to the National Institute for Infectious Diseases ‘Lazzaro Spallanzani’ (Rome, Italy) between 2005 and 2006. The research protocol was approved by the Institutional Review Board of the Hospital. Participants had given written informed consent before enrollment into the study. Clinical data were provided for each patient ().
Table 1. Clinical features of the nine HIV-infected patients recruited in this study
Faecal samples were collected, fixed and filtrated by using a 10% formalin disposable system (Meridian Biosciences, Cincinnati, OH, USA). Cryptosporidium oocysts were detected by a modified Ziehl–Neelsen staining (Becton Dickinson, Franklin Lakes, NJ, USA). After microscopic examination, the residual stool aliquot was stored at 4°C in formalin before DNA extraction.
DNA Extraction from Formalin-fixed Stools
DNA extraction was performed by the following modification of the QIAamp DNA stool mini kit (Qiagen, Hilden, Germany). Briefly, 200 μl of formalin-fixed stool samples were washed three times with phosphate-buffered saline (pH 7·4) and centrifuged to remove traces of formalin before DNA extraction. Samples were submitted to 15 freeze–thaw cycles (freezing in liquid nitrogen for 1 minute and heating at 65°C for 1 minute) and 1·4 ml of ASL buffer (QIAamp DNA stool mini kit; Qiagen) was added. The suspension was mixed, incubated for 5 minutes at 95°C and processed according to the manufacturer’s instructions (QIAamp DNA stool mini kit; Qiagen). DNA was eluted in 50 μl of H2O, previously heated at 65°C for 15 minutes. As positive control, DNA was extracted with the same procedure from 107 Percoll purified oocysts from the C. parvum isolate ISSC162, subgenotype IIaA15G2, kindly provided by the Department of Infectious, Parasitic and Immunomediated Diseases (Istituto Superiore di Sanità, Rome, Italy).
GP60-based Subgenotyping
A fragment of the GP60 gene was amplified using a previously described nested-PCR protocol (Sulaiman et al., Citation2005). Both PCR reactions, set up to a 25 μl volume, were performed as previously described (Glaberman et al., Citation2002), except for the annealing temperature (50 and 51°C, for first and second rounds, respectively). An aliquot of 5 μl from the primary PCR reaction was used as template in the secondary PCR reaction. Between primary and secondary PCRs, a purification step, performed by using a PCR clean-up system (Wizard; Promega, Fitchburg, WI, USA), was included to remove residual formalin affecting the secondary PCR. The resulting DNA fragments, loaded on 2·2% agarose gels (Sigma Aldrich, Milan, Italy), were visualized by Gel Doc XR System and analysed using the software Quantity One 4·6·3 (Bio-Rad, Hercules, CA, USA). The size of the amplified PCR products was estimated by comparison with the Flash Gel 50 bp–1·5 kb DNA ladder (Lonza, Rockland, ME, USA). Sequencing reactions were performed by using 1·6 pmol of inner GP60 primers (Sulaiman et al., Citation2005) and 1·5 μl of amplified DNA in a final volume of 10 μl, hence cycled according to the manufacturer (version 3·1; Applied Biosystems, Foster City, CA, USA). Consensus sequences corrected by base calling were probed against non-redundant GenBank databases using BLASTN algorithm (http://www.ncbi.nlm.nih.gov/BLAST/). The highest score matching was considered the most likely correct identification, and only an identity score of more than 98% was used as a threshold to be considered significant. GP60 subgenotypes were classified according to the nomenclature proposed by Sulaiman (Sulaiman et al., Citation2005). Nucleotide sequences were deposited into GenBank under accession nos. HM371366 to HM371374.
RESULTS
Clinical Manifestations of Cryptosporidiosis
Cryptosporidium was identified in stools from all nine patients, but in bronchoalveolar lavage (BAL), sputum and duodenal biopsy from three different patients. All patients but one had CD4 counts less than 50 cell/µl and a CDC C3, and were therefore susceptible to severe cryptosporidiosis. None of the subjects had suppressed HIV viraemia, with viral load ranging from 1·938 to more than 500 000 cp/ml, mainly (8/9 patients) due to poor compliance or failure of previous HAART regimens. Eight out of the nine Cryptosporidium cases were diagnosed between the moist months December and March, and one in September. Diarrhoea was complained of by all patients, and was watery for 8 of 9. Weight loss was reported by eight patients, with three of them experiencing a strong weight loss (∼15 kg in 1 month) and four patients suffering from wasting syndrome. Other common symptoms were: abdominal pain (7/9 patients), fever and vomiting (6/9 patients), and cough, often productive (4/9 patients); however, Cryptosporidium was isolated from sputum and BAL from only two patients. One patient had frank pneumonia and another had diffuse interstitial infiltrates. Nutritional biomarkers were also altered. Plasmatic albumin was low in 6/9 patients, suggesting strong and lasting malabsorption. As a consequence, the majority of patients (5/9 patients) required the administration of parenteral nutrition (PN) to contrast clinical condition worsening. Chronic diarrhoea and malabsorption led to electrolyte disturbances, especially hypokalaemia, except for two patients. The fecal occult blood resulted positive in six patients, but no patient clearly showed haematic stools. For patient 1, the histological examination of duodenal biopsy revealed sphere shapes suggestive of duodenal cryptosporidiosis. Also the patient 9 had symptoms compatible with biliary involvement, confirmed also by imagining evidences, but neither endoscopic retrograde cholecystopancreatography nor biopsies of biliary ducts were performed to certainly diagnose biliary cryptosporidiosis. Candidiasis was a recurrent co-infection, observed in 7/9 patients, mark of mucosal changes or altered intestinal immunity. We employed symptom features to categorize patients into three different groups: severe, moderate and mild cryptosporidiosis.
In the first set, patients 1, 3, 8 and 9 were grouped (). Patient 1 was a hepatitis C virus co-infected subject who presented withwasting syndrome, abdominal pain and vomiting. During the hospitalization, an esophagogastroduodenoscopy with biopsy was performed and the histological examination revealed a duodenal cryptosporidiosis (data not shown). Stool examination revealed the presence of Cryptosporidium oocysts (). Treatment with metronidazole and paromomycin failed with progressive worsening of clinical conditions. However, after a few days, the patient refused additional therapies and was discharged against medical advice (). Patient 3 presented with mucocutaneous Kaposi’s sarcoma and wasting syndrome. The patient was on HAART failure. He presented with fever, diarrhoea, dysphagia and weight loss. After hospitalization PN, glucose, amino acids and electrolyte solutions were started, but clinical conditions progressively deteriorated and after 32 days of hospital stay the patient died (). Patient 8 was a hepatitis C virus co-infected subject presenting with wasting syndrome and atypical mycobacteriosis who began both HAART and antimycobacterial therapy, with further improvement and discharge (). Patient 9 presented with wasting syndrome, esophageal candidiasis and systemic CMV infection with recurrent symptoms of diarrhoea and vomiting. Multiple HAART regimens were undertaken but failed. During hospitalization, he presented abdominal pain with marked elevations of amylases, lipases and cholestatic indexes. Abdominal ultrasound and magnetic resonance cholangiography showed intra- and extrahepatic dilatation of the biliary tree, edematous pancreas and a gallbladder filled with dense bile (suggesting duodenal cryptosporidiosis), but a duodenal biopsy was not undertaken. A chest X-ray evidenced diffuse interstitial infiltrates and pleural effusion. Sputum (data not shown) and stool examinations revealed Cryptosporidium oocysts (). His clinical conditions progressively worsened and he died.
Dark and pale grey apply to sets of patients referred as affected by severe and moderate cryptosporidiosis, respectively. Uncoloured patients are characterized by mild symptoms.
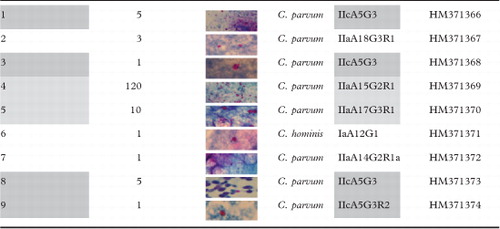
The second set (moderate symptoms) included patients 4 and 5 (). Patient 4 complained lack of appetite and had diarrhoea for about 2 months. Due to diarrhoea persistence, patient underwent a rectosigmoidoscopy suggesting ulcerative colitis; hence, a therapy with mesalazine and metronidazole was begun. Despite the therapy, the diarrhoea persisted and the patient began to additionally present productive cough and fever. A chest X-ray showed an interstitial reticulonodular pattern with a basal parenchymal consolidation (). Pulmonary tuberculosis was suspected but the BAL revealed the presence of Cryptosporidium oocysts (data not shown), also detected in large amounts in the stools (). Patient received PN, antibiotic therapy (paromomycin/azithromycin) and HAART with progressive improvement and further discharge. Patient 5 was admitted presenting fever, abdominal pain, diarrhoea and ingravescent loss of weight. Therefore, HAART was begun. Stool examination showed numerous Cryptosporidium oocysts () and a therapy with paromomycin/azithromycin was hence administrated. Afterwards, the clinical manifestations ameliorated and the patient was discharged.
The third set (mild symptoms) included patients 2, 6 and 7. Patient 2 complained watery diarrhoea, abdominal pain and loss of weight for 2 months and productive cough for 1 month. Stool examinations revealed Cryptosporidium oocysts (). HAART was not started; however, amelioration of clinical conditions was observed leading to his discharge. Patient 6 complained fever, watery diarrhoea, abdominal pain and dysphagia. Stool examinations revealed Cryptosporidium oocysts (). After a few days, diarrhoea diminished and the patient was discharged against medical advice. For patient 7, the seropositivity status was unknown on admission. He presented diarrhoea, dyspepsia and progressive loss of weight. Rifaximin was administered without any improvement; hence, the patient was hospitalized and therefore tested HIV-seropositive. Stool examination was positive for both Ascaris lumbricoides and Cryptosporidium (). Diarrhoea persisted despite the use of albendazole, but was reduced after paromomycin/azithromycin administration, leading to patient discharge.
DNA Extraction and Amplification from Formalin-fixed Stools
The optimization of the DNA extraction step from aged formalin-fixed stools was based on repeated phosphate-buffered saline washings for formalin excess removal followed by a thermal stress-induced rupture of oocysts for DNA release, by repeated freeze/thaw cycles and heating of the samples at 95°C. Mechanical oocyst disruption was not associated to an improved DNA yield and competence for amplification (data not shown). DNA competence was improved by choosing a small size DNA target (358 bp), thus minimizing the effect of formalin-induced DNA fragmentation.
GP60-based Subgenotyping
Nested-PCR at the GP60 locus yielded single products of the expected size for all samples. All products were successfully sequenced. BLAST analysis revealed that four isolates (samples 1, 3, 8 and 9) belonged to the C. parvum allelic family IIc: precisely, three samples (1, 3 and 8) were classified as subgenotype IIcA5G3 and one sample (9) as IIcA5G3R2 (). Four isolates, belonging to the allelic family IIa, were classified as IIaA18G3R1 (sample 2), IIaA15G2R1 (sample 4), IIaA17G3R1 (sample 5) and IIaA14G2R1a (sample 7) (). A single isolate (sample 6) belonged to the C. hominis subgenotype IaA12G1 ().
DISCUSSION
Based on symptom features, we categorized patients into three different groups: severe, moderate and mild cryptosporidiosis (). Consistently, the GP60 subgenotyping was discussed for the three patient sets. GP60 subgenotyping, for the first group of patients, identified only the IIcA5G3 lineage, while for the other two groups, a wider variety of lineages was reported. The C. parvum allelic family IIc has been frequently recorded (Jex and Gasser, Citation2010) and described from at least 12 countries: Australia (Jex et al., Citation2007; Jex et al., Citation2008; O’Brien et al., Citation2008), Guatemala (Peng et al., Citation2001), Japan (Abe et al., Citation2006), Kuwait (Sulaiman et al., Citation2005), Malawi (Peng et al., Citation2003), Perù (Cama et al., Citation2008), Portugal (Peng et al., Citation2001; Alves et al., Citation2006), South Africa (Leav et al., Citation2002), Spain (Jex and Gasser, Citation2008), The Netherlands (Wielinga et al., Citation2008), Slovenia (Soba and Logar, Citation2008) and Uganda (Akiyoshi et al., Citation2006). However, to date, this allelic family has been recorded almost exclusively in humans (Xiao and Feng, Citation2008; Bouzid et al., Citation2010) and only recently in hedgehogs (Dyachenko et al., Citation2010). Based on the clinical evidence, wasting syndrome appears strongly linked to infection with parasite of the family IIc (4/4 patients), whereas no wasting syndrome was observed in patients infected with Ia and IIa families (). Remarkably, for 2/4 IIcA5G3 patients (1 and 9), esophagogastroduodenoscopy and magnetic resonance cholangiography exams suggested an extended and atypical colonization (Lumadue et al., Citation1998) of the subtype IIcA5G3 to other sites (bile-duct/duodenum tract), different from sigmoid colon and rectum (Velásquez et al., Citation2010). The infection of the biliary tree represents a reservoir from which intestinal cryptosporidiosis may relapse and allows the organism to avoid luminal antiparasitic agents such as paromomycin (Chalmers and Davies, Citation2010). In these cases, drugs like nitaxozanide with biliary excretion should be used (Baishanbo et al., Citation2006). Indeed, the administration of paromomycin, even if integrated by metronidazole, failed as specific treatment of cryptosporidiosis (Palmieri et al., Citation2005; Pozio and Morales, Citation2005) for patient 1. For the second patient set (4 and 5), subtypes IIaA15G2R1 and IIaA17G3R1 were observed. The IIa allelic family comprises some 50 subgenotypes, has been detected in 26 countries worldwide, including Italy, with a prevalence of 25·5% in humans and of 57·8% in all other hosts (Jex and Gasser, Citation2010), and has been described as the only or prevalent family in some extended geographical areas (Plutzer and Karanis, 2007). The IIaA15G2R1 subtype, prevalently reported from farm animals, has zoonotic potential (Jex and Gasser, Citation2010). However, IIaA15G2R1 has been detected in human samples from Australia (O’Brien et al., Citation2008) and from sporadic human cases in Portugal (Alves et al., Citation2003), Belgium (Geurden et al., Citation2009), Ireland (Zintl et al., Citation2009), Kuwait (Sulaiman et al., Citation2005), Canada (Trotz–Williams et al., Citation2006), Japan (Amer et al., Citation2010) and the USA (Peng et al., Citation2003). Remarkably, this subtype was also linked to an outbreak associated with an open farm in Wales (UK) (Chalmers et al., Citation2005), supporting a zoonotic transmission. The other subtype IIaA17G3R1 was described both in calves and humans in Northern Ireland (Glaberman et al., Citation2002; Thompson et al., Citation2007), and in humans in Australia (Waldron et al., Citation2009).
Interestingly, an extra-intestinal cryptosporidiosis was noted for patients 4, similarly to patient 9. However, for patient 4, after receiving PN, the paromomycin/azithromycin combined therapy gradually improved the clinical conditions leading to patient discharge. The same symptoms were also seen in patient 5, despite the absence of pulmonary involvement. Only stool examination showed numerous Cryptosporidium oocysts; the same antibiotic therapy, used for patient 4, induced amelioration of her clinical manifestations.
For the third set of patients (2, 6 and 7), subtypes IIaA18G3R1, IIaA14G2R1a and IaA12G1 were observed. The first subtype has been very commonly reported in humans and cattle in Ireland (Zintl et al., Citation2009) and Australia (Waldron et al., Citation2009), and in patient 2, it self-resolved upon patient’s discharge. To the best of our knowledge, the IIaA14G2R1a subtype (), characterized in Germany for the first time by Broglia et al. (Citation2008) in cattle, is herein reported as a novel human subtype (patient 7). An important co-infection with A. lumbricoides was observed for this patient, but only a specific anti-Cryptosporidium treatment resolved diarrhoea. Interestingly, only for patient 6, a C. hominis subtype (IaA12G1) was reported. The allelic family Ia has a worldwide distribution and is significantly diverse at the subtype level (25 subtypes reported; Jex and Gasser, Citation2010). There may be slight differences in the clinical manifestations depending on C. hominis subtypes, as previously described (Cama et al., Citation2007). Apparently, the C. hominis infection for patient 6 was associated with the absence of fecal occult blood and required PN; there was no wasting syndrome and his weight loss was not remarkable when compared to the other patients.
Fixatives are essential for preservation of stool specimens. Buffered formalin is a traditional stool fixative, still considered the ‘gold standard’ in parasitology by virtue of its excellent long term preservative activity on intestinal parasites. However, the extraction of high-quality genomic DNA and its amplification is usually hampered by the high nucleic acid fragmentation chemically induced and by the presence of remnants of formalin that inhibit the amplification reaction. Because of these difficulties, we have developed a new protocol that yields DNA suitable for amplification of small size targets, which are less affected by sporadic DNA breakages. Cryptosporidium DNA extraction was performed directly from stool samples, without prior oocyst purification or concentration steps. The method relies on a chemical lysis sustained by a heating effect acting at two protocol stages before lysis and spin-column steps. However, no mechanical oocyst disruption, based on additional required equipments, was introduced to enhance the DNA yield, as previously reported (Zhu et al., Citation1998). The heating disruption appeared a resolutive determinant in the breakage of formalinized oocysts/sporozoites and in the release of amplifiable DNA. The protocol was introduced as a convenient bench-side procedure for stool specimen analysis and it was employed to select appropriate subgenotyping markers and to link subgenotyping lineages to individual manifestations of cryptosporidiosis.
Cryptosporidium subgenotyping is an important tool for the understanding its population structure. Given the need to investigate the genetic diversity of Cryptosporidium clinical isolates, still underestimated for technical difficulties to handle formalin-fixed stools, the method herein described may offer advantages for routine diagnosis and clinical epidemiological studies, despite referring to a small-scale patient dataset. Therefore, further evidence from larger sample size appears necessary to prove a statistically significant correlation between symptom severity and genotype. The interactionship may therefore provide insights on subgenotype lineages and HIV-associated cryptosporidiosis to unveil, on large patient cohorts, determinants of clinical diversity of cryptosporidiosis associated with genetic complexity of the parasite.
We would like to thank as source of support a grant from Ricerca Corrente (RC 2010) Children’s Hospital and Research Institute ‘Bambino Gesù’ to Lorenza Putignani.
We wish to thank Dr Fabio Tosini (Istituto Superiore di Sanità, Rome, Italy) for providing purified C. parvum oocysts, and Dr Andreina Santoro for her careful revision of the manuscript.
REFERENCES
- Abe N, Matsubayashi M, Kimata I, Iseki M. (2006). Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitology Research, 99, 303–305.
- Abubakar S, Aliyu H, Arumugam C, Hunter PR, Usman NK. (2007). Prevention and treatment of cryptosporidiosis in immunocompromised patients. The Cochrane Database of Systematic Reviews, (1), CD004932.
- Akiyoshi DE, Tumwine JK, Bakeera-Kitaka S, Tzipori S. (2006). Subtype analysis of Cryptosporidium isolates from children in Uganda. The Journal of Parasitology, 92, 1097–1100.
- Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. (2003). Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. Journal of Clinical Microbiology, 41, 2744–2747.
- Alves M, Xiao L, Antunes F, Matos O. (2006). Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitology Research, 99, 287–292.
- Amadi B, Mwiya M, Musuku J, Watuka A, Sianongo S, Ayoub A, Kelly P. (2002). Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet, 360, 1375–1380.
- Amer S, Matsubara R, Murakoshi F, Nakai Y. (2010). Molecular analysis of Cryptosporidium parvum HNJ-1 isolated in Japan. The Journal of Veterinary Medical Science, 72, 1647–1649.
- Baishanbo A, Gargala G, Duclos C, François A, Rossignol JF, Ballet JJ, Favennec L. (2006). Efficacy of nitazoxanide and paromomycin in biliary tract cryptosporidiosis in an immunosuppressed gerbil model. The Journal of Antimicrobial Chemotherapy, 57, 353–355.
- Bouzid M, Tyler KM, Christen R, Chalmers RM, Elwin K, Hunter PR. (2010). Multi-locus analysis of human infective Cryptosporidium species and subtypes using ten novel genetic loci. BMC Microbiology, 10, 213.
- Broglia A, Reckinger S, Cacciò SM, Nockler K. (2008). Distribution of Cryptosporidium parvum subtypes in calves in Germany. Veterinary Parasitology, 154, 8–13.
- Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, Vivar A, Ticona E, Navincopa M, Williamson J, Ortega Y, Gilman RH, Bern C, Xiao L. (2007). Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV. Journal of Infectious Diseases, 196, 684–691.
- Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L. (2008). Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerging Infectious Diseases, 14, 1567–1574.
- Certad G, Arenas-Pinto A, Pocaterra L, Ferrara G, Castro J, Bello A, Núñez L. (2005). Cryptosporidiosis in HIV-infected Venezuelan adults is strongly associated with acute or chronic diarrhea. The American Journal of Tropical Medicine and Hygiene, 73, 54–57.
- Chalmers RM, Ferguson C, Cacciò SM, Gasser RB, Abs El-Osta YG, Heijnen L, Xiao L, Elwin K, Hadfield S, Sinclair M, Stevens M. (2005). Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. International Journal for Parasitology, 35, 397–410.
- Chalmers RM, Davies AP. (2010). Minireview: clinical cryptosporidiosis. Experimental Parasitology, 124, 138–146.
- Dyachenko V, Kuhnert Y, Schmaeschke R, Etzold M, Pantchev N, Daugschies A. (2010). Occurrence and molecular characterization of Cryptosporidium spp. genotypes in European hedgehogs (Erinaceus europaeus L.) in Germany. Parasitology, 137, 205–216.
- Fall A, Thompson RC, Hobbs RP, Morgan-Ryan UJ. (2003). Morphology is not a reliable tool for delineating species within Cryptosporidium. Parasitology, 89, 399–402.
- Fayer R, Santín M, Macarisin D. (2010). Cryptosporidium ubiquitum n. sp. in animals and humans. Veterinary Parasitology, 172, 23–32.
- Feng X, Rich SM, Akiyoshi D, Tumwine JK, Kekitiinwa A, Nabukeera N, Tzipori S, Widmer G. (2000). Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Applied and Environmental Microbiology, 66, 3344–3349.
- Geurden T, Levecke B, Cacció SM, Visser A, de Groote G, Casaert S, Vercruysse J, Claerebout E. (2009). Multilocus genotyping of Cryptosporidium and Giardia in non-outbreak related cases of diarrhoea in human patients in Belgium. Parasitology, 136, 1161–1168.
- Glaberman S, Moore JE, Lowery CJ, Chalmers R M, Sulaiman I, Elwin K, Rooney PJ, Millar BC, Dooley JS, Lal AA, Xiao L. (2002). Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerging Infectious Diseases, 8, 631–633.
- Huang DB, Chappell C, Okhuysen PC. (2004). Cryptosporidiosis in children. Seminars in Pediatric Infectious Disease, 15, 253–259.
- Jex AR, Ryan UM, Ng J, Campbell BE, Xiao L, Stevens M, Gasser RB. (2007). Specific and genotypic identification of Cryptosporidium from a broad range of host species by nonisotopic single-strand conformation polymorphism (SSCP) analysis of nuclear ribosomal DNA. Electrophoresis, 28, 2818–2825.
- Jex AR, Gasser RB. (2008). Analysis of the genetic diversity within Cryptosporidium hominis and Cryptosporidium parvum from imported and autochtonous cases of human cryptosporidiosis by mutation scanning. Electrophoresis, 29, 4119–4129.
- Jex AR, Pangasa A, Campbell BE, Whipp M, Hogg G, Sinclair MI, Stevens M, Gasser RB. (2008). Classification of Cryptosporidium species from patients with sporadic cryptosporidiosis by use of sequence-based multilocus analysis following mutation scanning. Journal of Clinical Microbiology, 46, 2252–2262.
- Jex AR, Gasser RB. (2010). Genetic richness and diversity in Cryptosporidium hominis and C. parvum reveals major knowledge gaps and a need for the application of “next generation” technologies research review. Biotechnology Advances, 28, 17–26.
- Keating JP. (2005). Chronic diarrhea. Pediatrics in Review, 26, 5–14.
- Leav BA, Mackay MR, Anyanwu A, O’Connor RM, Cevallos AM, Kindra G, Rollins NC, Bennish ML, Nelson RG, Ward HD. (2002). Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infection and Immunity, 70, 3881–3890.
- Lumadue JA, Manabe YC, Moore RD, Belitsos PC, Sears CL, Clark DP. (1998). A clinicopathologic analysis of AIDS-related cryptosporidiosis. AIDS, 12, 2459–2466.
- Morgan UM, Xiao L, Fayer R, Lal AA, Thompson RC. (1999). Variation in Cryptosporidium: towards a taxonomic revision of the genus. International Journal for Parasitology, 29, 1733–1751.
- O’Brien E, McInnes L, Ryan UM. (2008). Cryptosporidium GP60 genotypes from humans and domesticated animals in Australia, North America and Europe. Experimental Parasitology, 118, 118–121.
- Palmieri F, Cicalini S, Froio N, Rizzi EB, Goletti D, Festa A, Macrí G, Petrosillo N. (2005). Pulmonary cryptosporidiosis in an AIDS patient: successful treatment with paromomycin plus azithromycin. International Journal of STD & AIDS, 16, 515–517.
- Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, Bern C, Sulaiman IM, Glaberman S, Lal AA, Xiao L. (2001). A comparison of Cryptosporidium subgenotypes from several geographic regions. The Journal of Eukaryotic Microbiology, 48 (Suppl.), 28S–31S.
- Peng MM, Meshnick SR, Cunliffe NA, Thindwa BD, Hart CA, Broadhead RL, Xiao L. (2003). Molecular epidemiology of cryptosporidiosis in children in Malawi. The Journal of Eukaryotic Microbiology, 50, 557–559.
- Plutzer J, Karanis P. (2009). Genetic polymorphism in Cryptosporidium species: an update. Veterinary Parasitology, 165, 187–199.
- Pozio E, Morales MAG. (2005). The impact of HIV-protease inhibitors on opportunistic parasites. Trends in Parasitology, 21, 58–63.
- Putignani L, Menichella D. (2010). Global distribution, public health and clinical impact of the protozoan pathogen Cryptosporidium. Interdisciplinary Perspectives on Infectious Diseases, 2010, 753512.
- Riggs MW. (2002). Recent advances in cryptosporidiosis: the immune response. Microbes and Infection, 4, 1067–1080.
- Robinson G, Wright S, Elwin K, Hadfield SJ, Katzer F, Bartley PM, Hunter PR, Nath M, Innes EA, Chalmers RM. (2010). Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): morphology, biology and phylogeny. International Journal of Parasitology, 40, 1539–1548.
- Soba B, Logar J. (2008). Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology, 135, 1263–1270.
- Strong WB, Gut J, Nelson RG. (2000). Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infection and Immunity, 68, 4117–4134.
- Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L. (2005). Unique endemicity of cryptosporidiosis in children in Kuwait. Journal of Clinical Microbiology, 43, 2805–2809.
- Thompson HP, Dooley JS, Kenny J, McCoy M, Lowery CJ, Moore JE, Xiao L. (2007). Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitology Research, 100, 619–624.
- Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine AS, Martin SW, Nydam DV, Jamieson F, Xiao L. (2006). Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitology Research, 99, 346–352.
- Velásquez JN, di Risio C, Marta E, Astudillo OG, Etchart C, Cucher MA, Chertcoff AV, Perissé E, Carnevale S. (2010). Occurrence of bile-duct/duodenal abnormalities in nine AIDS patients co-infected with Cryptosporidium hominis and/or C. parvum. Annals of Tropical Medicine and Parasitology, 104, 257–263.
- Waldron LS, Ferrari BC, Power ML. (2009). Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Experimental Parasitology, 122, 124–127.
- Wielinga PR, de Vries A, van der Goot TH, Mank T, Mars MH, Kortbeek LM, van der Giessen JW. (2008). Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. International Journal for Parasitology, 38, 809–817.
- Xiao L, Fayer R, Ryan UM, Upton SJ. (2004). Cryptosporidium taxonomy: recent advances and implications for public health. Clinical Microbiology Reviews, 17, 72–97.
- Xiao L, Ryan UM. (2004). Cryptosporidiosis: an update in molecular epidemiology. Current Opinion in Infectious Diseases, 17, 483–490.
- Xiao L, Feng Y. (2008). Zoonotic cryptosporidiosis. FEMS Immunology and Medical Microbiology, 52, 309–323.
- Zhu G, Marchewka MJ, Ennis JG, Keithly JS. (1998). Direct isolation of DNA from patient stools for polymerase chain reaction detection of Cryptosporidium parvum. The Journal of Infectious Diseases, 177, 1443–1446.
- Zintl A, Proctor AF, Read C, Dewaal T, Shanaghy N, Fanning S, Mulcahy G. (2009). The prevalence of Cryptosporidium species and subtypes in human faecal samples in Ireland. Epidemiology and Infection, 137, 1–8.