Abstract
We describe a case of a chronic myeloid leukemia patient displaying the chimeric BCR-ABL1 gene on 12p11. Chromosome analysis revealed complex chromosome aberration involving chromosomes 9, 12, and 22. Fluorescence in situ hybridization revealed an unusual signal pattern revealing the BCR-ABL1 fusion signal on chromosome 12, while no reciprocal ABL1-BCR fusion was detected on der(9) chromosome. The relocation of BCR-ABL1 fusion sequences to 12p11 site in our patient represents a rare type of variant translocation, as in almost all patients the chimeric BCR-ABL1 gene is located on der(22) chromosome. Our case illustrates the challenge of recognizing a complex pattern of cytogenetic aberrations that occur with variant t(9;22) and may add further information about clinical significance of unusual variant Ph rearrangements in CML patients receiving tyrosine kinase inhibitor treatment.
Introduction
Chronic myeloid leukemia (CML) is a clonal malignant disorder of a pluripotent hematopoetic stem cell characterized by the presence of Philadelphia chromosome (Ph) translocation t(9;22)(q34;q11). The result of the translocation is the juxtaposition of the 3′ portion of the Abelson oncogene (ABL1) from 9q34 to the 5′ portion of the breakpoint cluster region (BCR) on 22q11 leading to the formation of the BCR-ABL1 fusion gene resulting in the production of an abnormal tyrosine kinase protein.Citation1 Instead of the classical t(9;22) translocation, about 5% of patients have simple or complex variants of this translocation, involving one or more chromosomes in addition to 9 and 22.Citation2–Citation6
We report here a case of a CML patient carrying a complex variant Ph translocation resulting in relocation of BCR-ABL1 fusion sequences to chromosome 12. The variant location of BCR-ABL1 fusion sequences in a complex t(9;22;12) was not previously described and could be not detected without combination of conventional cytogenetics and molecular cytogenetic techniques. Our case demonstrates the importance of precise molecular cytogenetic characterization of variant Ph translocations, particularly in instances where several translocation partners occur.
Materials and methods
Chromosome preparation and fluorescence in situ hybridization analysis
Chromosome preparation and karyotyping were performed according to standard methods. Direct and 24-hour bone marrow cultures were established and chromosomes were G-banded with a Giemsa stain. Karyotype was described according to the International System for Human Cytogenetic Nomenclature.Citation7
Fluorescence in situ hybridization studies (FISH) were performed on slides prepared for cytogenetic analysis using a t(9;22) specific BCR-ABL1 dual color, dual fusion, and BCR-ABL1 tricolor, dual fusion + SpectrumAqua (extending from a point centromeric to the ASS gene and ends beyond the first exon (1b) of the ABL1 gene) probes directly labeled with SpectrumGreen, SpectrumOrange, and SpectrumAqua fluorochromes, respectively (Abbott Molecular, Vysis, IL, USA). To confirm the juxtaposition of 12p, dual color FISH was carried out with AML1 and TEL probes (Abbott Molecular), hybridizing to chromosome 21q22 and 12p13 regions. A minimum of 100 nuclei and a minimum of five metaphases were scored using a fluorescence microscope (Axioplan 2, Zeiss, Jena, Germany). Images were captured using a black and white CCD camera processed with the ISIS software program (MetaSystems, Altlussheim, Germany).
Molecular studies
Molecular studies were carried out from total RNA, extracted from the diagnostic bone marrow sample using the Trizol LS reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer's recommendations. cDNA was prepared from 2 µg of total RNA using Superscript II RT (Invitrogen) according to the manufacturer's instructions.
Results
A 26-year-old man with 1-month history of weight loss and abdominal pain was presented in September 2009. Physical examination revealed marked hepatosplenomegaly. The following haematological parameters were present: hemoglobin 11.8 g/l; platelets 945 × 109/l and white blood cells (WBC) 525 × 109/l with 45% neutrophils, 5% lymphocytes, 2% monocytes, 3% basophils, 20% myelocytes, 20% metamyelocytes, 2% promyelocytes, and blasts 3%). Bone marrow was markedly hypercellular with increased megakaryocytes and reduced erythroid lineage with dysplastic features leading to a diagnosis of CML. Serum biochemistry was remarkable for lactate dehydrogenase 1190 U/l (normal 100–190).
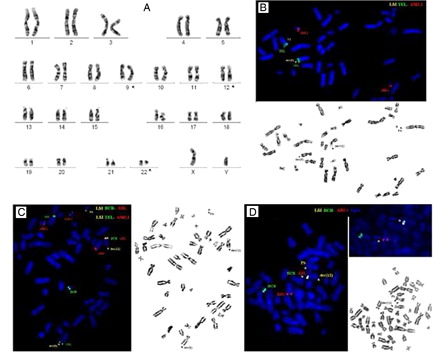
Chromosome analysis performed from diagnostic bone marrow sample revealed complex chromosome aberration involving chromosomes 9, 12, and 22 leading to a three-way chromosomal rearrangement: 46,XY,t(9;22;12)(q34;q11;p11) in all of 20 metaphases examined (A). The juxtaposition of chromosomal segment s from 12p to chromosome 9 was confirmed using the dual color TEL-AML1 probe revealing the translocation of TEL from the 12p13 site to 9q34 (green signal) (B and 1C). FISH with dual color BCR-ABL1 dual fusion probe revealed the fusion BCR-ABL1 signal in 90% of interphase cells. Hybridization on metaphases showed an unusual signal pattern, revealing the BCR-ABL1 fusion signal on chromosome 12 instead of the Philadelphia chromosome (C). Hybridization with LSI tricolor, dual fusion BCR-ABL1 translocation probe with the SpectrumAqua 9q34 probe confirmed the location of the BCR-ABL1 signal incorporating the ASS gene on chromosome 12, while no reciprocal ABL1-BCR fusion was detected on der(9) chromosome (D). Reverse transcriptase-polymerase chain reaction confirmed the presence of the BCR-ABL1 fusion showing the major BCR (MBCR) transcript.
Figure 1. G-banded metaphase from the diagnostic bone marrow showing the complex karyotype 46,XY,t(9;22;12)(q34;q11;p11) (A). FISH with the dual color, TEL-AML1 probe on metaphases showed juxtaposition of TEL from 12p13 to chromosome 9 generating signal (green signal) on der(9) (B). Dual Color FISH probe for BCR-ABL revealed variant location of BCR-ABL1 fusion, revealing the fusion BCR-ABL1 signal on a der(12) instead of a der(22) (C and D).
Therapy with hydroxyurea and imanitib (400 mg/day) was administrated. One month later, karyotyping revealed no cytogenetic response showing the t(9;22;12) rearrangement in all of 20 metaphases examined. He continued his treatment with imatinib; however, no cytogenetic response was achieved during the course of disease. In May 2010, karyotyping revealed t(9;22;12) in 20 examined metaphases and FISH studies showed 90% BCR-ABL1-positive cells. In July 2010, he discontinued imatinib therapy for a period of 1 month leading to a rapid increase of blood counts to 145 × 109/l; however, after restarting imatinib therapy (400 mg/day) he was well until December 2010, when his WBC increased to 147 × 109 (56% neutrophils, 1% lymphocytes, 2% monocytes, 2% basophils, 12% myelocytes, 22% metamyelocytes, 2%, blasts 1%, and platelets 1083 × 109/l) associated with hepatosplenomegaly. The patient traveled outside for further treatment.
Discussion
The common molecular event underlying classical, cryptic, and variant Philadelphia translocations is the constitutive activation of the ABL1 tyrosine kinase on 9q34, considered to be the initial pathologic event in CML.Citation1–Citation4,Citation8 Since tyrosine kinase activity is essential for the transforming activity of the BCR-ABL1 fusion protein, a specific small molecule tyrosine kinase inhibitor such as imatinib, could be an effective treatment for patients with CML. However; relocation of BCR-ABL1 fusion sequences is an infrequent event and its phenotypic consequence in patients receiving tyrosine kinase inhibitor therapy remains to be investigated. In the most recent analysis by Marzocchi et al.Citation9 including 559 CML patients, in almost all the cases, the BCR-ABL1 fusion sequence was located on der(22) and relocation of BCR-ABL1 sequences was detected only in 2 patients.
The Mitelman database for chromosomal aberrations in cancer was searched for cases carrying a complex variant Ph translocation involving both chromosomes 9 and 22 as well as the short arm of chromosome 12.Citation10 Because breakpoints on 12p may be difficult to ascertain, we included cases with 12p11–13 bands. A review of the literature showed four cases with t(9;22;12)Citation2,Citation11–Citation13 and two cases with t(9;12;22).Citation14,Citation15 Chromosomal breakpoint 12p11 observed in our patient was detected in two of these cases.Citation13,Citation14 In additional three cases, 12p was involved in variant Ph translocations with rearrangements of four and seven chromosomes, respectively, indicating that the short arm of chromosome 12 may be part of more complex multistep events in CML.Citation3,Citation16
Rearrangements of the short arm of chromosome 12 are common in a broad spectrum of hematological malignancies, including CML, where they usually occur as part of a more complex karyotype. These rearrangements are frequently accompanied by concomitant submicroscopic deletions, variously mapped to all three bands on the short arm of chromosome 12.Citation17 Thus, we can speculate that the deletion encompassing the reciprocal ABL1-BCR product from the derivative chromosome 9 in our patient might be accompanied by concomitant loss of genomic material from 12p as a potential side effect of DNA breakage event at 12p.
While the variant Ph translocation involving 9q34, 12p11–13, and 22q11 bands has been described in several cases, this is a first report in the literature to the best of our knowledge with a detailed molecular cytogenetic characterization disclosing the location of BCR-ABL1 fusion sequences. Metaphase FISH analysis alone, allowed us to precisely characterize this rare Ph variant that cannot be detected by conventional cytogenetic or interphase FISH analysis.
Karyotype and FISH results suggest a two-step mechanism in our patient, i.e. that the standard t(9;22) is formed followed by a second translocation involving derivative chromosomes 9 and 22 and chromosome 12 with simultaneous deletion of ABL1-BCR fusion sequences from chromosome der(9) chromosome. Despite of the potential curative role of tyrosine kinase inhibitors in CML, only transient hematological response was achieved in our patient and noteworthy, imatinib discontinuation for a short period of time resulted in a rapid hematological relapse indicating that multistep events in CML might be markers of genomic instability.
Conclusion
The formation mechanism of variant location of BCR-ABL1 fusion, the fusion sequence with leukemogenic potential, has remained unexplored. Because of the small number of such cases, their association with clinical outcome in imatinib era remains to be clarified.Citation18
References
- Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–53.
- Gorusu M, Benn P, Li Z, Fang M. On the genesis and prognosis of variant translocations in chronic myeloid leukemia. Cancer Genet Cytogenet. 2007;173:97–106.
- Bennour A, Sennana H, Laatiri MA, Elloumi M, Khelif A, Saad A. Molecular cytogenetic characterization of variant Philadelphia translocations in chronic myeloid leukemia: genesis and deletion of derivative chromosome 9. Cancer Genet Cytogenet. 2009;194:30–7.
- Sessarego M, Fugazza G, Bruzzone R, Ballestrero A, Miglino M, Bacigalupo A. Complex chromosome rearrangements may locate the bcr/abl fusion gene sites other than 22q11. Haematologica. 2000;85:35–9.
- Bennour A, Bellâaj H, Ben Youssef Y, Elloumi M, Khelif A, Saad A, et al. Molecular cytogenetic characterization of Philadelphia-negative rearrangements in chronic myeloid leukemia patients. J Cancer Res Clin Oncol. 2011;137:1329–36.
- Bakshi SR, Patel BP, Brahmbhatt MM, Trivedi PJ, Gajjar SB, Iyer RR, et al. Complex karyotype with a masked Philadelphia translocation and variant BCR-ABL fusion in CML. Cancer Genet Cytogenet. 2009;189:142–3.
- Lisa G, Shaffer, Slovak ML, Campbell LJ. ISCN 2009: an international system for human cytogenetic nomenclature. S. Karger, AG, 2009.
- Zagaria A, Anelli L, Albano F, Storlazzi CT, Liso A, Roberti MG, et al. A fluorescence in situ hybridization study of complex t(9;22) in two chronic myelocytic leukemia cases with a masked Philadelphia chromosome. Cancer Genet Cytogenet. 2004;150:81–5.
- Marzocchi G, Castagnetti F, Luatti S, Baldazzi C, Stacchini M, Gugliotta G, et al. Variant Philadelphia translocations: molecular-cytogenetic characterization and prognostic influence on frontline imatinib therapy, a GIMEMA Working Party on CML analysis. Blood. 2011;117:6793–800.
- Mitelman F, Johansson B, Martens F, (eds.). Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (2012). http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107:76–94.
- Nishigaki H, Inazawa J, Misawa S, Nishida K, Okuda T, Horiike S, et al. Distribution of breakpoint within the breakpoint cluster region (bcr) in chronic myelogenous leukemia with a complex Philadelphia chromosome translocation. Am J Hematol. 1989;32:194–9.
- Ohyashiki K, Ohyashiki JH, Kinniburgh AJ, Rowe J, Miller KB, Raza A, et al. Transposition of breakpoint cluster region (3′ bcr) in CML cells with variant Philadelphia translocations. Cancer Genet Cytogenet. 1987;26:105–15.
- Dow LW, Raimondi SC, Culbert SJ, Ochs J, Kennedy W, Pinkel DP. Response to alpha-interferon in children with Philadelphia chromosome-positive chronic myelocytic leukemia. Cancer. 1991;68:1678–84.
- Storlazzi CT, Specchia G, Anelli L, Albano F, Pastore D, Zagaria A, et al. Breakpoint characterization of der(9) deletions in chronic myeloid leukemia patients. Genes Chromosomes Cancer. 2002;35:271–6.
- El-Zimaity MM, Kantarjian H, Talpaz M, O'Brien S, Giles F, Garcia-Manero G, et al. Results of imatinib mesylate therapy in chronic myelogenous leukaemia with variant Philadelphia chromosome. Br J Haematol. 2004;125:187–95.
- Sato Y, Kobayashi H, Suto Y, Olney HJ, Davis EM, Super HG, et al. Chromosomal instability in chromosome band 12p13: multiple breaks leading to complex rearrangements including cytogenetically undetectable sub-clones. Leukemia. 2001;15:1193–202.
- Stagno F, Viagneri P, del Fabro V, Stella S, Cupri A, Massimino M, et al. Influence of complex variant chromosomal translocations in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors. Acta Oncologica. 2010;49:506–8.