Abstract
Aim
To assess the prognostic role of myeloid transcription factor gene CEBPA (CCAAT/enhancer binding protein-α), a novel gene involved in leukemia in Egyptian adults AML.
Materials and Methods
Screening for CEBPA mutations was assessed using PCR-single-strand conformation polymorphism (PCR-SSCP) in pretreatment bone marrow samples from 55 newly diagnosed adult AML.
Results
CEBPA mutations were found in 11 (20%) of 55 AML patients. They had significantly higher hemoglobin (P = 0.037), and lower LDH (P = 0.003) levels when compared to those without. CEBPA mutations were frequently detected in M4 (45.5%) and M2 (27.2%) subtypes, and significantly associated with normal karyotype (90.9%, P = 0.007). We distinguished six cases with two different mutations or one homozygous mutation (CEBPAdouble-mut) as well as five cases with only one single heterozygous mutation (CEBPAsingle-mut). Patients with CEBPA mutations had significantly higher complete remission (P = 0.047), lower mortality (p = 0.047). Double CEBPA mutant cases showed longer disease free survival (DFS) and overall survival (OS) when compared to wild type CEBPA (for DFS; median = 27 versus 24 months respectively; P = 0.009 and for OS; median = 28 versus 25 months respectively; p = 0.008). No significant differences were found between CEBPAsingle-mut cases and wild type cases regarding DFS and OS (for DFS; median = 13 versus 24 months respectively; P = 0.615 and for OS; median = 14 versus 25 months respectively; P = 0.703).
Conclusion
CEBPA mutation status is known to be a prognostic factor for favorable outcome in AML patients. CEBPAdouble-mut is associated with favorable DFS and OS. In contrast, CEBPAsingle-mut AMLs survival studies did not differ significantly with wild-type cases. These results demonstrate significant underlying heterogeneity within CEBPA mutation positive AML with prognostic relevance. Based on these findings, we propose that CEBPAdouble-mut should be clearly defined from CEBPAsingle-mut AML and considered as a separate entity in the classification of AML. Furthermore, incorporation of CEBPA mutation status into novel risk-adapted therapeutic strategies in Egypt will improve the currently disappointing cure rate of this group of patients.
Introduction
Acute myeloid leukemia (AML) is clinically and molecularly heterogeneous disease. Currently, cytogenetic findings provide the most important prognostic information and are used to guide risk-adapted treatment strategies.Citation1 The identification of molecular markers that precisely differentiate a patient's risk could improve treatment outcome,Citation2 and deeply refine the prognosis of patients with AML.Citation3
Transcription factor CCAAT/enhancer binding protein α (CEBPA), encoded by CEBPA gene that is located at chromosome 19q13.1,Citation4 was originally isolated from rat liver that bound to viral enhancer sequences CCAAT.Citation5 CEBPA is the original member of the basic region leucine zipper (bZIP) class of transcription factors.Citation6 CEBPA protein consists of two N-terminal transactivation domains, a basic DNA-binding region, and a C-terminal leucine zipper domain.Citation7 Wild-type (WT) CEBPA exists in two translational isoforms, p42 and p30.Citation8
It has been shown that CEBPA is specially expressed in human myelomonocytic cell lines and regulates the expression of several granulocyte-specific genes,Citation9 and is involved in the regulation of myelopoiesis.Citation10 CEBPA transcriptionally activates the promotors of the myeloid-specific receptors for macrophage colony-stimulating factor (M-CSF), granulocyte colony-stimulating factor (G-CSF), and granulocyte-macrophage colony-stimulating factor (GM-CSF) in myeloid cells.Citation11 Finally, CEBPA-deficient mice exhibit a blockage in the granulocytic differentiation at the myeloblast level with a complete absence of mature granulocytes occurring early in the fetal development, whereas other blood cell types are present in normal proportions.Citation12
CEBPA mutations affect either the N-terminal part of the protein that specifically abolish translation of full-length (42 kDa) CEBPA, leading to overexpression of a shorter (30 kDa) form or the bZIP domain that affects dimerization and DNA binding.Citation13
Genetic abnormalities in AML frequently occur in transcriptional control elements, leading to uncontrolled proliferation and/or differentiation arrest. Recently, mutations of the gene coding for CEBPA leading to inactivation of its transcriptional potential have been reported in hematologic malignancies with a particular incidence of 7–10% in AML cells.Citation14 It has been speculated that CEBPA mutations might contribute to the differentiation block specific to AML.Citation15 Interestingly, the presence of a CEBPA mutation was associated with significantly better clinical outcome.Citation16 However, others found that CEBPA mutation was not of prognostic importance in their AML patients.Citation17 In the current study, we investigated a group of AML patients for CEBPA mutation to clarify its biological and prognostic impact on disease outcome.
Materials and methods
Patients and samples
The present study included 55 newly diagnosed de novo adult AML patients representing various French-American-British (FAB) subtypes. They were 25 males and 30 females with mean age (45.65 ± 16 years). They were selected from patients admitted to the Mansoura Oncology Center during 2009–2011. A written informed consent was obtained from the patients prior to their enrollment in this study.
Patients were diagnosed according to standard diagnostic methods including cytomorphological, cytochemical, immunophenotypic (positivity by flowcytometry was defined as an expression in at least 20% of cells in the gated population of interest, compared to internal negative control cellsCitation18), and cytogenetic evaluation.Citation19 In addition, 10 healthy subjects with matched age and sex were selected to act as a control group. Bone marrow (BM) samples from patients with AML were subjected to Ficoll-Hypaque (Pharmacia LKB, Uppsala, Sweden) density gradient centrifugation. All samples taken at diagnosis were confirmed to contain more than 90% leukemia cells after enrichment by centrifugation. Inclusion criteria were newly diagnosed AML patients, with no previous treatment. Exclusion criteria were history of exposure to chemotherapy/radiotherapy, and secondary AML patients, markedly impaired hepatic or renal functions, concurrent severe and/or uncontrolled medical conditions (e.g. uncontrolled diabetes, infection, hypertension, etc.), family history of hematological malignancies, and FLT-3 ITD gene mutations positive cases by polymerase chain reaction (PCR).
Cytogenetic risk classification
Cytogenetic risk classification was done according to Revised Medical Research Council prognostic classification;Citation20 favorable risk group defined by the presence of t(15;17) (q22;q21), t(8;21) (q22;q22), inv(16)(p13;q22), unfavorable risk group defined by the presence of 11q23 abnormalities (del(ll)(q23), t(4;ll)(q21;q23), t(10;ll)(pl2;q23), and t(6;ll)(q27;q23). Entities not classified as favorable or adverse were identified as intermediate risk group.
Treatment protocol
Patients aged less than 60 years received the standard ‘3 + 7’ induction chemotherapy protocol: doxorubicin (30 mg/m2 per day) for 3 days and cytarabine (100 mg/m2 per day as a continuous 24 hours intravenous infusion) for 7 days.Citation21 Patients aged more than 60 years with performance status 0–2 and minimal comorbidity received standard therapy with 3 + 7; however, those with performance status more than 2 who are unfit for therapy were treated with low dose Ara-c 20 mg/m2/12 hours for 10 days every month. Patients with acute promyelocytic leukemia (M3) who received All-trans retinoic acid plus anthracycline.Citation22 BM aspiration was done between 21 and 28 days after initiation of chemotherapy. Consolidation comprised of three to four courses of high-dose cytosine arabinoside (3 g/m2 every 12 hours on Days 1, 3, and 5; total, 18 g/m2). Following this, patients were followed up once every 3 months with clinical examination and complete blood counts. A BM aspiration was done if there was any suggestion of a relapse on clinical examination or peripheral smear.
Methods
All patients and control samples were analyzed for mutation of the CEBPA gene using genomic PCR–SSCP (single-strand conformation polymorphism) method.Citation23
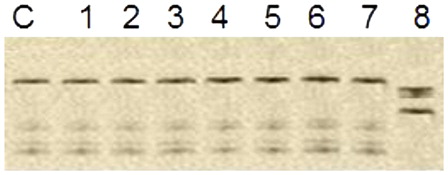
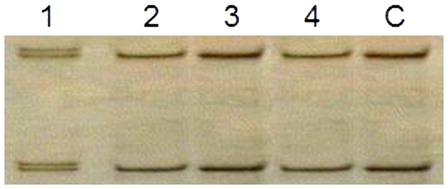
Genomic DNAs were extracted from frozen BM mononuclear cells collected at diagnosis by using a DNA extraction kit (Puregene Gentra System, Minneapolis, MN, USA) according to the manufacturer's instructions. Two overlapping primer pairs () were used to amplify the entire coding region of human CEBPA. PCR reactions were run in a final volume of 50 µl containing genomic DNA (100 ng), KCl (100 nmol/l), Tris-HCl (30 mmol/l, pH 8.05), MgCl2 (5 mmol/l), 5 volume % DMSO, primers (12.5 pmol/l of each), nucleotides (400 µmol/l of each), and Taq DNA polymerase (0.05 U/μl, Qiagen, Hilden, Germany). The mixture was denatured at 94°C for 1 minute, annealed at 61°C for 40 seconds, and extended at 72°C for 90 seconds for 35 cycles, with a final step for 10 minutes at 72°C. SSCP was performed on PCR products, that were mixed with 10 volumes of loading buffer, denatured at 96°C, quenched on ice immediately, and applied to 5% polyacrylamide gel electrophoresis. Normal gene exhibits a specific conformational pattern, while a mutant gene displays a pattern with a different electrophoretic mobility (mobility shift).
Table 1. Primers used to amplify CEBPA from genomic DNA
The electrophoretic conditions were optimized by doing some considerations; adding glycerol to the polyacrylamide gel that lowers the pH of the electrophoresis buffer more specifically, which can detect single base substitutions in 600–800 bp fragments with high sensitivity;Citation24 electrophoresis temperature was constant to obtain best results. The conditions for sample application suggested by the manufacturer were done. To identify candidate oncogenic alleles, any changes that were also present in normal control samples were excluded.
Criteria of response and survival definitions
Complete remission (CR) required a BM blasts <5.0%; absence of blasts with Auer rods; absence of extramedullary disease; absolute neutrophil count > 1.0 × 109/l; platelet count > 100 × 109/l; with independence of red cell transfusions. Relapse was defined as BM blasts ≥5.0%; or reappearance of blasts in the blood; or development of extramedullary disease.Citation25 Resistant disease (RD) was defined as more than 15.0% BM blasts after induction therapy. Overall survival (OS) was defined as the time from entry to death. For patients achieving first CR, disease-free survival (DFS) was defined as the time from first CR to an event (death in CR or relapse).
Statistical analysis
Data analysis was performed using SPSS for window version16.0 (SPSS, Inc., Chicago, IL, USA). Differences between two groups were done by t-test and Chi-square test. Survival studies were done using Kaplan–Meier curve and Log rank test.
Results
Mutation frequency of CEBPA in AML samples
CEBPA mutations were detected in 11 of 55 AML patients (20%) at initial diagnosis. All healthy control subjects had WT allele.
We distinguished six cases with two different mutations or one homozygous mutation (CEBPAdouble-mut) as well as five cases with only one single heterozygous mutation (CEBPAsingle-mut) ( and ).
Clinicohematological characteristics of AML patients with CEBPA mutation
Patients with CEBPA mutation had significantly higher hemoglobin (P = 0.037), and lower lactate dehydrogenase (LDH) levels (P = 0.003) than those without the mutation. On the other hand, no significant differences were observed in most pre-treatment clinical characteristics between patients with wild and mutant CEBPA with respect to age (P = 0.939), gender (P = 0.176), platelet count (P = 0.457), peripheral, and BM blast cells (P = 0.284, P =0.785) ().
Table 2. Hematological characteristics of AML patients with CEBPA mutation
All cases were divided according to FAB classification; 2 cases had M0, 4 cases had M1, 11 cases had M2, 4 cases had M3, 18 cases had M4, 12 cases had M5, and 4 cases had M6 ().
Patients with CEBPA mutations had higher frequencies of FAB AML M4 (45.5%) and M2 (27.2%) subtypes, with no significant difference between mutated and wild CEBPA groups regarding different FAB subtypes ().
The leukemic cells from all CEBPA-mutated patients in the present study, expressed the CD33 antigens; in addition, the majority of CEBPA-mutated leukemic cells expressed cMPO (90.9%), CD13 (81.8%), CD117 (81.8%), while CD14 had lesser incidence (54.5%). HLA-DR and CD11b had the least incidence (45.5% for both) in CEBPA-mutated patients ().
CEBPA mutation and cytogenetics
Cytogenetic data were available for all patients, 30 patients were of normal karyotype, 7 had t(8;21)(q22;q22), 7 had inv16(p13.1q22), 4 had t(15;17)(q22;q12), 2 had t(6;9)(p23;q34), 5 had 11q23 abnormalities (1 had del(ll)(q23), 2 had t(4;ll)(q21;q23), 1 had t(10;ll)(pl2;q23), and 1 had t(6;ll)(q27;q23)).
All cases of CEBPA mutations were NK-AML except for one case with inv16, with significant difference between mutated and non-mutated CEBPA groups regarding cytogenetics (P = 0.007).10 (33.3%) of 30 patients with intermediate risk and 1 (5.6%) of 18 patients with favorable risk cytogenetics had CEBPA mutations. On the other hand, none of those with unfavorable risk cytogenetics had CEBPA mutations, with no significant difference between mutated and non-mutated CEBPA groups regarding cytogenetic risk (P = 0.179) ().
Table 3. Cytogenetics of AML patients with CEBPA mutation
CEBPA mutation and clinical outcome
Thirty-six AML cases achieved CR (65.5%), 10 cases were refractory (18.2%), 4 cases relapsed after CR (7.3%) and 19 cases died (9 cases died during induction therapy, 7 cases were refractory, 1 cases died after relapse, and 2 cases died after achieving CR).
The course of the disease in patients with CEBPA mutation was favorable, a total of 36 (65.5%) of the 55 AML patients achieved CR with significantly higher CR (P = 0.047), and lower mortality (P = 0.047) in patients with CEBPA mutation. However, no statistical difference in relapse and refractoriness between both groups (P = 0.299, P = 0.382) ().
Table 4. Clinical outcome in relation to CEBPA gene mutation status
Table 5. Survival in CEBPA mutations versus non-CEBPA mutation in AML patients
All six patients with CEBPA double mutations reached CR (100%), while CR was achieved in four out of the five (80%) patients with CEBPA single mutations, as one patient had RD.
Survival studies demonstrated that CEBPA mutation predicted significantly longer DFS (median = 26 versus 21 months for mutant versus wild respectively; P = 0.005). This good prognosis associated with CEBPA mutations was also observed in terms of OS (median = 27 versus 25 months for mutant versus wild type respectively; P = 0.013) ().
CEBPAdouble-mut cases showed longer DFS and OS when compared to WT CEBPA (for DFS; median = 27 versus 24 months, respectively; P = 0.009 and for OS; median = 28 versus 25 months, respectively; P = 0.008) ( and ).
Figure 3. Kaplan–Meier estimates of DFS. DFS among CEBPAdouble-mut versus CEBPA WT AML (P = 0.009) and versus CEBPAsingle-mut AML (P = 0.163); CEBPAsingle-mut AML versus CEBPAwt AML (P = 0.634; pooled = 0.024).
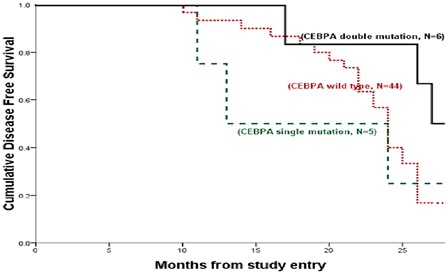
Figure 4. Kaplan–Meier estimates of OS. OS among CEBPAdouble-mut versus CEBPAwt AML (P = 0.008) and versus CEBPAsingle-mut AML (P = 0.197); CEBPAsingle-mut AML versus CEBPAwt AML (P = 0.712; pooled = 0.021).
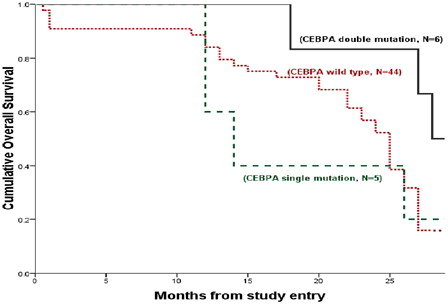
No significant differences were found between CEBPAsingle-mut cases and WT cases regarding DFS and OS (for DFS; median = 13 versus 24 months, respectively; P = 0.615 and for OS; median = 14 versus 25 months, respectively; P = 0.703).
Discussion
The growth inhibitory and differentiation promoting function of CEBPA make it an attractive candidate for tumor suppression. It is likely that impairment of either of these may induce leukemia. In the last few years, various mechanisms have been suggested through which CEBPA is negatively regulated in AML.Citation26 We tested this by screening the CEBPA locus in 55 samples from AML patients.
We used SSCP to detect CEBPA mutations under optimal conditions; approximately 80–90% of the potential base exchanges are detectable by SSCP.Citation27
Mutations of CEBPA gene were found in 11 (20%) of 55 AML patients, these results confirm the previously reported incidence of CEBPA mutation in AML (18–20%).Citation17,Citation28,Citation29 Others reported lower incidence of CEBPA mutations.Citation29–Citation32 This difference may be due to variable number of cases, and also some studies focused on NK-AML. In addition, the sensitivity of the method used affects the results. Analysis of larger control populations will be required for the detection of rare polymorphisms.
No significant differences were found between WT CEBPA versus mutant CEBPA groups as regard to age and sex, in accordance with other studies.Citation29,Citation33 However, others found that younger age was associated with a greater number of mutations.Citation34
The present study showed no significant differences regarding total leukocyte count (TLC), platelets count, as well as blast cells percentages in peripheral blood and in BM between mutated and non-mutated groups. These results agreed with previous reports.Citation29,Citation33 On the other hand, significant lower platelets count and higher blast cells percentage were reported by others.Citation35,Citation36
Hemoglobin level was higher, while LDH value was lower in patients with CEBPA mutation compared to patients without (P = 0.037 and P = 0.003, respectively). The present study agreed with others who reported significant higher hemoglobin levelsCitation15,Citation34 and lower LDH levels.Citation27,Citation33 However, others reported no significant difference in LDH level.Citation15 This discrepancy in results may be related to the relatively limited number of patients included in the present study.
Regarding FAB repartition, mutated CEBPA gene showed highest incidence in M4 and M2, respectively. On the other hand, no mutations were noticed in M0 and M3. Many studies reported highest CEBPA mutation in M1 and M2.Citation37,Citation38 M4, M5, and M6 were also reported.Citation37,Citation38 The higher incidence of CEBPA mutations in M1, M2, and M4 FAB subtypes supports a critical role of CEBPA gene function in the intermediate stages of granulocytic differentiation.Citation39
No statistical differences between patients with mutated CEBPA and those without mutation regarding cMPO, CD13, CD33, CD117, HLA-DR CD14, and CD11b antigens. Previous immunophenotyping studies revealed that CEBPA mutation was associated with HLA-DR + CD7 + CD15 + CD34+ cells in AML.Citation32 Others reported more mature blast phenotype, (positive for CD33 and CD11c).Citation40
Our study reported CEBPA mutation in 10 (33.3%) of 30 NK-AML and in only 1(4%) of 25 AML with abnormal cytogenetics. Lin et al.,Citation36 from Taiwan detected CEBPA mutations in 35% of those with normal karyotype. These frequencies were higher than those reported in the western countries (15%) with normal karyotype.Citation13,Citation15,Citation41 This may be related to the variable number of patients included in each study, ethnicity, genetic, as well as geographic heterogeneity of AML. In addition to the type and accuracy of the methods used for the detection of CEBPA mutation. Others mentioned also the heterogeneity of CEBPA mutations as a cause of variation in CEBPA mutation incidence.Citation42
The frequency of CR was significantly higher (P = 0.047) and mortality was lower (P = 0.047) in patients with CEBPA mutations than in patients with WT. Also, DFS and OS were significantly longer (median 26 and 27 months, P = 0.005, P = 0.013, respectively) for patients with CEBPA mutations. Similar results were also obtained in other studies that linked CEBPA mutations with a favorable outcome in AML.Citation15,Citation29,Citation43 However, others found that CEBPA mutation was not of prognostic importance in AML patients.Citation17
In AMLs with two CEBPA mutations, the mutations are typically on different alleles.Citation44 Hence, in these cases, no WT CEBPA protein is expressed. A similar condition is found in AMLs carrying a homozygous CEBPA mutation.Citation31 However, there are also AMLs that only show one single heterozygous mutation and thus retain expression of a WT allele.Citation39,Citation44
In line with previous data,Citation31 DFS and OS were significantly better for CEBPAdouble-mut cases compared with cases with WT CEBPA (for DFS; median = 27 versus 24 months, respectively; P = 0.009 and for OS; median = 28 versus 25 months, respectively; P = 0.008). As well as no significant differences were found between CEBPAsingle-mut cases and WT cases regarding DFS and OS (for DFS; median = 13 versus 24 months, respectively; P = 0.615 and for OS; median = 14 versus 25 months, respectively; P = 0.703).
It is unclear as to why CEBPAdouble-mut AMLs would have a better outcome than those with a single heterozygous mutation. One explanation could be that a single-mutant CEBPA allele is not sufficient for leukemogenesis and requires cooperating mutations, which may be in CEBPA itself or in other genes. Of note, recent data indicate that germline CEBPA mutations predispose to AML, and the acquisition of a second, somatic CEBPA mutation may then contribute to AML development.Citation45
Other genomic abnormalities may accompany CEBPA mutations and have a prognostic impact in the CEBPA-mutated patients.Citation46 Further prospective studies of the genetic and protein alterations in more patients are needed to clarify this point.
It could be concluded that the molecular assessment of this factor at diagnosis offers valuable additional prognostic information and thereby will markedly affect therapeutic decisions in Egyptian population with AML. CEBPA mutant AML should at least be distinguished according to the presence of CEBPAdouble-mut and CEBPAsingle-mut. Our findings indicate that there is relevant prognostic heterogeneity within AML patients with CEBPA mutations. Double CEBPA mutations are associated with more favorable prognosis. However, the number of patients with CEBPA mutation is limited in our collection of patients, and larger series are needed to definitively assess its prognostic significance.
Acknowledgements
We thank the technicians for their excellent laboratory assistance. Laboratory work was performed using the logistics of the Center for Clinical Pathology, Faculty of Medicine, Mansoura University, Egypt. All the experiments were carried out in compliance with the current laws of Egypt.
References
- Byrd JC, Mrozek K, Dodge RK. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B. Blood. 2002;100:4325–36.
- Creutzig U, Kaspers GJ. Revised recommendations of the International Working Group for diagnosis, standardization of response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2004;22:3432–3.
- Renneville A, Boissel N, Gachard N, Naguib D, Bastard C, Botton S, et al. The favorable impact of CEBPA mutations in patients with acute myeloid leukemia is only observed in the absence of associated cytogenetic abnormalities and FLT3 internal duplication: clinical trials and observations. Blood. 2009;113:5090–3.
- Filip R, Dana D, Tomas J, Ivana J, Zlatuse K, Jiri M. CEBPA gene mutational status: a complete screening using high-resolution melt curve analysis. Mol Diagn Ther. 2009;13:195–200.
- Johnson PF, Landschulz WH, Graves BJ, McKnight SL. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987;1:133–46.
- Graves BJ, John PH, McKnight SL. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986;44:565–76.
- Tsukadaa J, Yoshidab Y, Kominatoc Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19.
- Hankey W, Silver M, Sun H, Zibello T, Berliner N, Khanna-Gupta A. Differential effects of sumoylation on the activities of CCAAT enhancer binding protein alpha (C/EBPα) p42 versus p30 may contribute in part, to aberrant C/EBPα activity in acute leukemias. Hematol Rep. 2011;3:13–7.
- Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–35.
- Sonda N, Chioda M, Zilio S, Simonato F, Bronte V. Transcription factors in myeloid-derived suppressor cell recruitment and function. Curr Opin Immunol. 2011;23:279–85.
- Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88(4):1234–47.
- Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc Natl Acad Sci USA. 1997;94:569–74.
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat Genet. 2001;27:263–70.
- Tenen DG. Abnormalities of the CEBPα transcription factor: a major target in acute myeloid leukemia. Leukemia. 2001;15:688–9.
- Frohling S, Schlenk RF, Stolze I, Bihlmayr J, Benner A, Kreitmeier S, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–33.
- Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CAJ, Wouters BJ, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117:2469–75.
- Snaddon J, Smith ML, Neat M, Cambal-Parrales M, Dixon-McIver A, Arch R, et al. Mutations of CEBPA in acute myeloid leukemia FAB types M1 and M2. Genes Chromosomes Cancer. 2003;37:72–8.
- Bain BJ, Barnett D, Linch D, Matutes E, Reilly JT. Revised guideline on immunophenotyping in acute leukaemias and chronic lymphoproliferative disorders. Clin Lab Haematol. 2002;24:1–13.
- Schoch C, Kern W, Schnittger S, Hiddemann W, Haferlach T. Karyotype is an independent prognostic parameter in therapy-related acute myeloid leukemia (t-AML): an analysis of 93 patients with t-AML in comparison to 1091 patients with de novo AML. Leukemia. 2004;18:120–5.
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65.
- Greer J, Baer M, Kinnery M. Acute myelogenous leukemia. In: , Lee G, Foester J, Leukens J, Pareskerva F, Greer J, Rogers G, (eds.) Wintrobe's clinical hematology, 12th edn. Philadelphia: Lippincott Williams and Wilkins; 2009:2045–62.
- Kanamaru A, Takemoto Y, Tanimoto M, Murakami H, Asou N, Kobayashi T, et al. All trans retinoic acid for the treatment of newly diagnosed acute promyelocytic leukemia. Japan Adult Leukemia Study Group. Blood. 1995;85:1202–6.
- Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–80.
- Kukita Y, Tahira T, Sommer S, Hayashi K. SSCP analysis of long DNA fragments in low pH gel. Hum Mutat. 1997;10:400–7.
- Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel. Blood. 2010;115:453–74.
- Trivedi AK, Pal P, Behre G, Singh SM. Multiple ways of CEBPA inhibition in myeloid leukaemia. Eur J Cancer. 2008;44:1516–23.
- Nollau P, Wagener C. Methods for detection of point mutations: performance and quality assessment. Clin Chem. 1997;43:1114–28.
- Bienz M, Ludwig M, Mueller BU, Leibundgut EO, Ratschiller D, Solenthaler M, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–24.
- Preudhomme C, Sagot C, Boissel N, Cayuela J, Tigaud I, Botton S, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood. 2002;100:2717–23.
- Schwieger M, Löhler J, Fischer M, Herwig U, Tenen DG, Stocking C. A dominant-negative mutant of C/EBPalpha, associated with acute myeloid leukemias, inhibits differentiation of myeloid and erythroid progenitors of man but not mouse. Blood. 2004;103:2744–52.
- Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–91.
- Kim S, Kim DH, Jang JH, Jung CW, Jang MA, Ki CS, et al. Novel mutations in CEBPA in Korean patients with acute myeloid leukemia with a normal karyotype. Ann Lab Med. 2012;32:153–7.
- El-Sharnouby JA, Ahmed LM, Taha AM, Kamal O. Prognostic significance of CEBPA mutations and BAALC expression in acute myeloid leukemia patients with normal karyotype. Egypt J Immunol. 2008;15:131–43.
- Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Orthod. 2010;28:2739–47.
- Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, et al. Prognostic significance of, and gene and MicroRNA expression signatures associated with, CEBPA mutations in ctogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. 2008;1(26):5078–87.
- Lin L, Chen C, Lin D, Tsay W, Tang J, Yeh Y, et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11:1372–9.
- Dohner MKH, Bloomfield CD. Influence of new molecular prognostic markers in patients with karyotypically normal acute myeloid leukemia: recent advances. Curr Opin Hematol. 2007;14:106–14.
- Schnittgera S, Bacherb U, Edera C, Lohsec P, Haferlacha C, Kerna W, et al. A copy number repeat polymorphism in the trans activation domain of the CEPBA gene is possibly associated with a protective effect against acquired CEBPA mutations: an analysis in 1135 patients with AML and 187 healthy controls. Exp Hematol. 2011;39:87–94.
- Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19:329–34.
- D'Alò F, Di Ruscio A, Guidi F, Fabiani E, Greco M, Rumi C, et al. PU.1 and CEBPA expression in acute myeloid leukemia. Leuk Res. 2008;32:1448–53.
- Gombart AF, Hofmann WK, Kawano S, Kawano S, Takeuchi S, Krug U, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein a in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–40.
- Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28:570–7.
- Barjesteh van Waalwijk van Doorn-Kho srovani S, Erpelinck C, Meijer J, Erpelinck C, Meijer J, van Oosterhoud S, et al.. Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4:31–40.
- Pabst T, Mueller BU. Transcriptional dysregulation during myeloid transformation in AML. Oncogene. 2007;26:6829–37.
- Pabst T, Eyholzer M, Haefliger S, Schardt J, Mueller BU. Somatic CEBPA mutations are a frequent second event in families with germline CEBPA mutations and familial acute myeloid leukemia. J Clin Oncol. 2008;26:5088–93.
- Perrotti D, Marcucci G, Caligiuri MA. Loss of C/EBPα and favorable prognosis of acute myeloid leukemias: a biological paradox. J Clin Oncol. 2004;22:582–4.