Abstract
The occurrence of resistance mutations in the Abl kinase domain plays a central role in drug treatment failure in chronic myeloid leukemia (CML) patients. Among them, the T315I mutation at the gatekeeper position affects a common Abl kinase contact residue and confers complete resistance to all known ATP-competitive BCR-ABL inhibitors. In the present study, an allele-specific oligonucleotide reverse transcriptase polymerase chain reaction assay was used to detect T315I mutation in a cohort of 60 imatinib-resistant CML patients. In terms of disease phase, 43 patients (71%) were in late chronic phase, 4 (7%) in accelerated phase, and 13 (22%) in blastic phase. The prevalence of the T315I mutation was found to be 7% (4/60). All four patients with mutation were in advance phases and had previously lost all their responses. The results of the study confirmed that this method is low cost and easy tool to operate for T315I mutation screening and direct sequencing should be performed in positive cases for confirmation.
Introduction
Chronic myeloid leukemia (CML) is a clonal disorder of multipotent hematopoietic progenitor cells, and is characterized by t (9; 22) (q34; q11) that yields the BCR-ABL fusion oncoprotein with constitutive tyrosine kinase activity.Citation1 CML cells undergo more changes and as time passes, it can convert to more aggressive phases of disease.
Given the central role of the BCR/ABL oncoprotein in CML pathogenesis, tyrosine kinase inhibitors (TKIs) have been introduced in the last decade. Imatinib mesylate, a TKI, has become as the standard in the treatment of CML patients.Citation2 Imatinib selectively induces cell growth inhibition and apoptosis of malignant cells harboring the oncoprotein.Citation3 Despite the success of imatinib therapy in the improvement of survival in CML, treatment failure remains a significant challenge and some patients ultimately develop resistance to imatinib treatment and undergo clinical relapse.Citation4
To date, more than 90 different amino acid substitutions in ABL tyrosine kinase domain have been identified.Citation5 Among them T315I, the mutation at the gatekeeper position, decreases the affinity of the TKIs for the tyrosine kinase domain by affecting a common Abl kinase contact residue and confers complete resistance to all known ATP-competitive BCR-ABL inhibitors (imatinib, dasatinib, and nilotinib).Citation6–Citation8 It also confers additional features to the leukemogenic potential of BCR/ABL.Citation9 Hence, T315I is of particular concern in clinical settings, and hematopoietic stem cell transplantation is the only established therapeutic option.Citation10 So, screening for this mutation is now recommended for all CML patients undergoing TKIs treatment and should be performed as early as possible to detect the lowest levels of the mutant clone.Citation11,Citation12
In this study, we used allele-specific oligonocleotide-reverse transcriptase polymerase chain reaction (ASO-RT-PCR) method to identify the T315I mutation in 60 Iranian patients with imatinib resistant. In addition, we evaluated the clinical findings in the patients with mutation during analysis.
Materials and methods
Patients
Over 350 CML patients are currently being treated with imatinib in the Hematology-Oncology and Stem Cell Transplantation Research Center of Shariati Hospital. Since 2009, 60 imatinib-resistant patients have been enrolled in the study according to hematological, cytogenetic, and molecular responses to identify T315I mutation based on European Leukemia Net (ELN) guidelines.Citation2,Citation13 Twenty-five out of the 60 patients (42%) were pretreated with hydroxyurea and/or interferon-α before imatinib, and the remaining 35 patients (58%) received imatinib as first-line therapy. These patients were in different phases of the disease and received treatment with high-dose imatinib (600–800 mg/day). An additional group of 10 healthy donors and 10 patients presenting with non-resistant CML served as a negative control group.
Quantitative qRT-PCR for BCR-ABL
Total RNA was isolated from peripheral blood using QIAzol Lysis Reagent (Qiagen, Hilden, Germany). First-strand cDNA was synthesized from 2 µg RNA using random hexamers and Moloney-murine-leukaemia virus reverse transcription reagents (Fermentas, Life Sciences, Vilnius, Lithuania) in total volume of 20 µl. Quantification of BCR-ABL fusion transcript and ABL control gene were performed using BCR-ABL Fusion Quant® Kit (Ipsogen, Marseille, France) according to a standardized protocol.Citation14 All standards and patient samples were run in duplicate. The copy number of BCR-ABL and ABL were calculated based on the standard curve. Finally, the BCR-ABL copy numbers were expressed as the ratios BCR-ABL/ABL in percent.
ASO-RT-PCR assay for detection of T315I mutation
For the ASO-RT-PCR, the reaction included three primers F1 (5′-cgc aac aag ccc act gtc t-3′) and R1 (5′-tcc act tcg tct gag ata ctg gat t-3′) based on reports by Branford et al.Citation15,Citation16 and Rm (5′-cgt agg tca tga act caa-3′) specific for T315I mutation a report by Willis et al.Citation17 These sets of PCR primers generates two PCR products: the mutant allele gave a specific amplification band of 304 bp along with an 863-bp fragment related to the entire ABL KD domain, while the wild ABL allele just gave an amplification band of 863 bp. The reaction was performed in thermal cycler (ABI, Foster City, CA, USA) in a reaction mixture containing 2 µl cDNA, 0.4 µM of each primer, 12.5 µl Taq DNA Polymerase Master Mix RED kit (Ampliqon, Copenhagen, Denmark). The thermal profile used for amplification was: initial denaturation of 94°C for 3 minutes followed by 35 cycles of denaturation at 94°C for 10 seconds, primer annealing at 57°C for 20 seconds, and extension at 72°C for 40 seconds. The final extension was at 72°C for 5 minutes. After amplification, 10 µl of PCR products was visualized under ultraviolet light by electrophoresis on a 2% (W/V) agarose gel stained with ethidium bromide.
Bidirectional sequencing
We used a semi-nested RT-PCR followed by bidirectional sequencing to detect T315I mutation for ASO-RT-PCR-positive patients according to Branford et al.Citation16 In brief, an 863-bp PCR product, which codes for amino acids 220–506 of the ABL protein, was purified using QIAquick PCR purification kit (Qiagen) and then sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Sequencing products were purified by the DyeEx 2.0 Spin kit (Qiagen). Bidirectional sequencing was carried out in an ABI Prism 3130 Genetic Analyzer (Applied Biosystems). Finally, the sequences were compared to the wild-type ABL sequence (GenBank accession number X16416).
Results
The clinical characteristics of the patients are summarized in . Mean age was 44 years (range: 12–77). Mean disease duration in these patients was 58 months (range: 12–192). In terms of disease phase, 43 patients (71%) were in late chronic phase, 4 (7%) in accelerated phase, and 13 (22%) in blastic phase.
Table 1. Characteristics of 60 CML patients with imatinib-resistant
The mean of BCR-ABL copy numbers in these patients was 47% (range: 10–258%). All patients did not achieve or lost a complete cytogenetic response and major molecular response. At the time of sampling, 37 Patients (62%) had no a hematological response (NHR), while 18 (30%) showed a hematological response (HR), and 5 (8%) showed a partial cytogenetic response (PCgR).
A shows the results of the ASO-RT-PCR amplification. In a mutated case, two fragments of 304 and 863-bp were seen. In non-mutated cases resistant to imatinib, non-resistant patients and healthy donors only the larger fragment was observed.
Figure 1. Patients with T315I mutation. (A) T315I mutation was detected by ASO-RT-PCR, as described. In brief, the ASO-RT-PCR protocol amplifies an 863-bp product (both mutant and wild-type alleles and serves as an internal control) and a 304-bp product (when the patient carries the T315I mutation). Lanes 1 and 2: patient A; lanes 3 and 4: patient B; lane 5: patient C; and lane 6: patient D. Lanes 7–10, 11, and 12 show patient with imatinib resistance without mutation, negative control and 100-bp size marker, respectively. (B) Direct sequencing analysis of samples A1, A2, B1, B2, C, and D are shown. As can be seen, the resistant mutated cells expanded from 20% (sample A1) to 90% (sample A2) in patient A and 50% (sample B1) to 60% (sample B2) in patient B. Patient C shows a mix of T315I mutation and wild-type ABL, each %50 and Patient D shows a mix of T315I mutation about %30 and wild-type ABL about %70 in individual samples available.
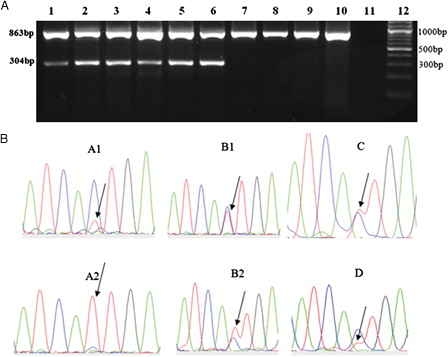
The prevalence of the T315I mutation was found to be 7% (4/60). Out of four patients with mutation who had lost all their responses, two were in accelerated phase and two were in blastic phase. A positive ASO-RT-PCR result for mutation was also confirmed by direct sequencing analysis showing the changes made by C1308 T where an ‘act’ coding for Threonine mutate to an ‘att’ coding for Isoleucine (T315I) in exon 7(B).
shows patient's details with detection of T315I mutation. Patient A who had two samples (A1 and A2) was a 54-year-old man with 25 months of disease duration and was in late chronic phase. The BCR-ABL copy numbers were %8.3. T315I-positive CML cells were confirmed by sequencing analysis showing a mix of T315I mutation about %20 as small peak and wild-type ABL about %80 (sample A1). Then, after 4 months progressed into the accelerated phase and T315I mutation and BCR-ABL copy numbers increased to %90 and %21.8, respectively (sample A2).
Table 2. Patient's details with detection of T315I mutation
Patient B who also had two samples (B1 and B2) was a 50-year-old woman with 118 months of disease duration and was in blastic phase. She was pretreated with hydroxyurea and interferon-α before treatment with imatinib at 400 mg daily. In the absence of response to imatinib at the standard dose (400 mg/day) and higher dose (600–800 mg/day), it was discontinued and replaced by nilotinib and dasatinib. The present study included samples collected during the course of dasatinib therapy. BCR-ABL copy numbers were %49 and T315I-positive CML cells were confirmed by sequencing analysis showing a mix of T315I mutation and wild-type ABL, each %50 (sample B1). After 3 months of treatment, the peak of T315I mutation and BCR-ABL copy numbers increased to %60 and %86.6, respectively (sample B2). Finally, the patient died due to blast crisis.
Patient C who was a 62-year-old female with 15 months of disease duration was in accelerated phase during treatment with imatinib 800 mg/day. The BCR-ABL copy numbers were %67.6 and T315I-positive CML cells were confirmed by sequencing analysis showing a mix of T315I mutation and wild-type ABL, each %50.
Finally, Patient D was a 12-year-old man with 36 months disease duration who developed a blastic phase after imatinib and desatinb therapy. Then, the patient underwent bone marrow transplantation (BMT), but unfortunately he relapsed after 3 months. The present study included post-BMT sample. The BCR-ABL copy numbers were %45 and T315I-positive CML cells were confirmed by sequencing analysis showing a mix of T315I mutation about %30 and wild-type ABL about %70. The patient was then followed-up, but unfortunately died due to blast crisis.
Discussion
The most common mechanism of imatinib resistance includes the acquisition of point mutations within the kinas domain. Mutations can affect the TKI and BCR-ABL tyrosine kinase interaction directly, as well as indirectly if their presence shifts the thermodynamic equilibrium from the inactive toward the active conformation of the enzyme.Citation18 The presence of a mutation at amino acid 315, where an ‘act’ coding for Threonine change to an ‘att’ coding for isoleucine (T315I), obliterates binding of imatinib and all currently available TKIs.Citation19,Citation20 Hence, identification of the T315I has prognostic significance and early detection of the mutation would enable to identify patients who may benefit from an allogeneic stem cell transplantation.Citation10,Citation14,Citation21
Among 60 imatinib-resistant patients, 7% had mutations. The mutation frequency previously reported ranges between 2% and 24% with variability related to the sensitivity of tests applied in different studies, as well as the higher percentage of patients with more advanced disease.Citation8,Citation17,Citation22–Citation26 Ethnic origin of patients and differences in pharmacokinetic profiles should also be considered.Citation24 The present study included a high proportion of patients in CP phase. Our patients with mutation had lost all responses and were in the accelerated phase and blastic phase indicating close relationship between the mutation and disease progression. On the other hand, according to studies, the emergence of mutant may be due to increase of clonal instability and the proliferation rate in advanced disease.Citation22,Citation26–Citation28
Compared to other techniques, ASO-RT-PCR provides a rapid and inexpensive method for detection of mutation. Furthermore, the assay is easy-to-perform and has high specificity and sensitivity. Therefore, it could be helpful for routine use in the clinical laboratory to screen BCR-ABL T315I point mutation. On the other hand, direct sequencing is recognized as a confirmation method for any screening tests, but high costs and low sensitivity (10–20%) make it inappropriate for routine clinical use.Citation29 Since this mutation confer additional features to the leukemogenic potential of BCR/ABL and is the only mutation that confers resistance against virtually all ATP competitors,Citation30 it is better to perform the T315I mutation assay with ASO-RT-PCR method in all patients with imatinib resistance. If the result is positive direct sequencing is recommended for confirmation.
We found that although the mutated clone was present at low levels in parallel with the lower BCR-ABL copy numbers at the time of initial evaluation, it is replaced over time and showed an increase in BCR-ABL copy numbers. Therefore, continuous molecular monitoring during imatinib therapy is an efficient method to follow response to treatment and can predict early stages in the development of imatinib resistance in patients with CML. Patients with treatment failure or suboptimal response to imatinib often demonstrate a steady trend of BCR-ABL level.Citation31 Hence, monitoring BCR-ABL transcript levels by qRT-PCR can specify the patients to evaluate mutation in case of significant increase in the number of copies.
In conclusion, the results of the study showed the advantages of ASO-RT-PCR assay to identify the T315I mutation in TKI-resistant CML patients for therapeutic decisions. In addition, it indicated that the T315I mutation in imatinib-resistant patients occurred in those with advanced phases or those with mutation eventually progressed to advanced phases.
Acknowledgments
This study was supported and performed in collaboration with the Department of Hematology, Faculty of Allied Medicine, Tehran University of Medical Sciences and the Hematology-Oncology and Stem Cell Transplantation Research Center in Shariati Hospital.
References
- Sherbenou DW, Druker BJ. Applying the discovery of the Philadelphia chromosome. J Clin Invest. 2007;117(8):2067–74.
- Baccarani M, Cortes J, Pane F, Niederwieser D, Saglio G, Apperley J, et al. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27(35):6041–51.
- Savage DG, Antman KH. Imatinib mesylate – a new oral targeted therapy. N Engl J Med. 2002;346(9):683–93.
- Shah NP. Loss of response to imatinib: mechanisms and management. Hematol Am Soc Hematol Educ Program. 2005:183–7.
- Soverini S, Hochhaus A, Nicolini FE, Gruber F, Lange T, Saglio G, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European LeukemiaNet. Blood 2011;118(5):1208–15.
- O'Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood 2007;110(7):2242–9.
- Soverini S, Iacobucci I, Baccarani M, Martinelli G. Targeted therapy and the T315I mutation in Philadelphia-positive leukemias. Haematologica 2007;92(4):437–9.
- Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: by the GIMEMA Working Party on Chronic Myeloid Leukemia. Clin Cancer Res. 2006;12(24):7374–9.
- Mian AA, Schull M, Zhao Z, Oancea C, Hundertmark A, Beissert T, et al. The gatekeeper mutation T315I confers resistance against small molecules by increasing or restoring the ABL-kinase activity accompanied by aberrant transphosphorylation of endogenous BCR, even in loss-of-function mutants of BCR/ABL. Leukemia 2009;23(9):1614–21.
- O'Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr Opin Genet Dev. 2006;16(1):92–9.
- Baccarani M, Castagnetti F, Gugliotta G, Palandri F, Soverini S. Response definitions and European Leukemianet Management recommendations. Best Pract Res Clin Haematol. 2009;22(3):331–41.
- Jones D, Kamel-Reid S, Bahler D, Dong H, Elenitoba-Johnson K, Press R, et al. Laboratory practice guidelines for detecting and reporting BCR-ABL drug resistance mutations in chronic myelogenous leukemia and acute lymphoblastic leukemia: a report of the Association for Molecular Pathology. J Mol Diagn. 2009;11(1):4–11.
- Baccarani M, Saglio G, Goldman J, Hochhaus A, Simonsson B, Appelbaum F, et al. Evolving concepts in the management of chronic myeloid leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 2006;108(6):1809–20.
- Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood 2006;108(1):28–37.
- Branford S, Rudzki Z, Parkinson I, Grigg A, Taylor K, Seymour JF, et al. Real-time quantitative PCR analysis can be used as a primary screen to identify patients with CML treated with imatinib who have BCR-ABL kinase domain mutations. Blood 2004;104(9):2926–32.
- Branford S, Rudzki Z, Walsh S, Grigg A, Arthur C, Taylor K, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood 2002;99(9):3472–5.
- Willis SG, Lange T, Demehri S, Otto S, Crossman L, Niederwieser D, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood 2005;106(6):2128–37.
- Redaelli S, Piazza R, Rostagno R, Magistroni V, Perini P, Marega M, et al. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J Clin Oncol. 2009;27(3):469–71.
- Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354(24):2531–41.
- Kantarjian H, Giles F, Wunderle L, Bhalla K, O'Brien S, Wassmann B, et al. Nilotinib in imatinib-resistant CML and philadelphia chromosome-positive ALL. N Engl J Med. 2006;354(24):2542–51.
- Giles FJ, Cortes J, Jones D, Bergstrom D, Kantarjian H, Freedman SJ. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood 2007;109(2):500–2.
- Nicolini FE, Corm S, Le QH, Sorel N, Hayette S, Bories D, et al. Mutation status and clinical outcome of 89 imatinib mesylate-resistant chronic myelogenous leukemia patients: a retrospective analysis from the French intergroup of CML (Fi(phi)-LMC GROUP). Leukemia 2006;20(6):1061–6.
- Qin Y, Chen S, Jiang B, Jiang Q, Jiang H, Li J, et al. Characteristics of BCR-ABL kinase domain point mutations in Chinese imatinib-resistant chronic myeloid leukemia patients. Ann Hematol. 2011;90(1):47–52.
- Markose P, Chendamarai E, Balasubramanian P, Velayudhan SR, Srivastava VM, Mathews V, et al. Spectrum of BCR-ABL kinase domain mutations in patients with chronic myeloid leukemia from India with suspected resistance to imatinib-mutations are rare and have different distributions. Leuk Lymphoma. 2009;50(12):2092–5.
- Kim SH, Kim D, Kim DW, Goh HG, Jang SE, Lee J, et al. Analysis of Bcr-Abl kinase domain mutations in Korean chronic myeloid leukaemia patients: poor clinical outcome of P-loop and T315I mutation is disease phase dependent. Hematol Oncol. 2009;27(4):190–7.
- Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood 2003;102(1):276–83.
- Soverini S, Martinelli G, Rosti G, Bassi S, Amabile M, Poerio A, et al. ABL mutations in late chronic phase chronic myeloid leukemia patients with up-front cytogenetic resistance to imatinib are associated with a greater likelihood of progression to blast crisis and shorter survival: a study by the GIMEMA Working Party on Chronic Myeloid Leukemia. J Clin Oncol. 2005;23(18):4100–9.
- Nicolini FE, Mauro MJ, Martinelli G, Kim DW, Soverini S, Muller MC, et al. Epidemiologic study on survival of chronic myeloid leukemia and Ph(+) acute lymphoblastic leukemia patients with BCR-ABL T315I mutation. Blood 2009;114(26):5271–8.
- Kang HY, Hwang JY, Kim SH, Goh HG, Kim M, Kim DW. Comparison of allele specific oligonucleotide-polymerase chain reaction and direct sequencing for high throughput screening of ABL kinase domain mutations in chronic myeloid leukemia resistant to imatinib. Haematologica 2006;91(5):659–62.
- Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood 2008;112(13):4808–17.
- Moravcova J, Zmekova V, Klamova H, Voglova J, Faber E, Michalova K, et al. Differences and similarities in kinetics of BCR-ABL transcript levels in CML patients treated with imatinib mesylate for chronic or accelerated disease phase. Leuk Res. 2004;28(4):415–9.