Abstract
Objectives
Human platelet-specific alloantigens (HPA) are polymorphic epitopes which vary among ethnic groups.
Background
In Tunisia, HPA frequencies were determined in North and centre; however, the pattern of HPA in South Tunisian population is not been studied yet. The aim of this work was to determine allelic frequencies of HPA-1, -3, and -5 systems in south Tunisian population, in order to estimate the risk of anti-platelet allo-immunization and to create a register of HPA-typed blood donors.
Methods
Our study concerned 212 unrelated healthy, regular blood donors from southern Tunisia. Allelic polymorphisms of each system were determined using a polymerase chain reaction with sequence-specific primers.
Results
Genotype frequencies a/a, a/b, and b/b were, respectively, 0.670, 0.288, and 0.042 for HPA-1 system, 0.430, 0.462, and 0.108 for HPA-3 system, and 0.750, 0.241, and 0.009 for HPA-5 system. The allele frequencies were 0.814 and 0.186 for HPA-1a and -1b alleles; 0.660 and 0.340 for HPA-3a and -3b alleles and 0.870, and 0.130 for HPA-5a and -5b alleles.
Discussion
The reported frequencies are more similar to those of Caucasians than those of north Tunisian population.
Introduction
Human platelet-specific alloantigens (HPA) are polymorphic epitopes of platelet membrane glycoproteins (GP).Citation1 To date, 24 platelet-specific alloantigens have been defined by immune sera, of which 12 are grouped into six bi-allelic systems (HPA-1, -2, -3, -4, -5, and -15). The molecular basis of 22 of the 24 serologically defined antigens has been identified. For 21 platelet allo-antigens, the difference between self and non-self is distinguished by a single amino acid substitution, caused by a single-nucleotide polymorphism (SNP) in the gene encoding the relevant GP membrane.Citation2
These epitopes are the target of platelet alloantibodies which can appear during pregnancy or after blood transfusion.Citation1 HPA allo-immunization is commonly responsible of three important clinical manifestations: Neonatal allo-immune thrombocytopenia (NAIT), post-transfusional purpura (PTP), and platelet transfusion refractoriness (PTR).Citation3,Citation4
The relative importance of each HPA system in anti-platelet allo-immunization varies among ethnic groups.Citation5–Citation7 Thus, the study of HPA polymorphism in different populations presents several interests. The first one is epidemiologically important in clinical practice and particularly the prediction of alloimmune disease.Citation8 The second one gives support allo-immunized patients by the determination of HPA phenotypes and anti-HPA antibodies in order to allow the diagnosis, establish a prognosis, and to provide compatible platelets when needed.Citation9,Citation10 Finally, these studies present a genetic anthropological interest.Citation8
Serological HPA phenotyping is very limited while platelet genotyping by molecular approach is widely recommended. Several techniques for SNP typing have been described for platelet genotyping. Polymerase chain reaction (PCR) amplification with sequence-specific primers (PCR-SSP) is currently used.Citation7
The aim of this work was to determinate the allelic frequencies of HPA-1, -3, and -5 systems, in order to estimate the risk of anti-platelet allo-immunization and to create a register of HPA-typed blood donors in the south of Tunisia.
Materials and methods
Blood donors, samples, and gene amplification
This study concerned 212 blood donors (148 men and 64 women). All of them are unrelated regular healthy volunteers, from southern Tunisia (Sfax City: n =172, Delegations of Sfax and South: n = 40). They were divided into 147 whole-blood donors and 65 platelets donors. Genomic DNA was extracted by the salting-out method,Citation11 from venous blood samples of 10 ml each collected on EDTA.
HPA-1, -3, and -5 systems were genotyped by PCR-SSP. The sequences of primers used here are according to Klüter et al.Citation7 A pair of primers amplifying a conserved region of the gene for C-reactive protein (CRP) was included in each tube, as internal control ().
Table 1. Sequences and length of designed primers for HPA-1, -3, -5 and the two internal positive control primers
PCR was performed as described previously by Klüter et al.Citation7 using a Gene Amp PCR System 9700, Applied Biosystem Thermal Cycler (Singapore), in a volume of 20 µl. Positive and negative controls were included in each series.
PCR products were visualized by UV trans-illumination after electrophoresis on agarose gel at 1–2% and staining with ethidium bromide.
Statistical analysis
Genotype frequencies were determined by direct counting. Allele frequencies were calculated from genotype frequencies. The confidence interval was set at 5%. The validity of Hardy–Weinberg equilibrium for each of the HPA system was tested by χ2 test.Citation12 Comparisons of allele frequencies between different populations were assessed by the χ2 test (reduced gap). The significance level was chosen to be 5%.
Results
In our study, PCR conditions were set different for HPA-1b, -3b, -5a, and -5b alleles. In fact, the alleles of the same system have different hybridization temperatures. Our work was therefore organized in several series of blood donors for each allele.
We have chosen nine HPA-specific primers and two CRP primers (internal positive control) for HPA-1, -3, and -5 systems genotyping. We used also DNAs with known HPA genotypes. In , we give an example of the results obtained by the PCR-SSP method showing that primers specifically amplify the desired platelet HPA-1a-related DNA and that the two alloantigen-specific primers clearly distinguish between the two alleles. The 440 bp amplification product of CRP control primer was present in all lanes. Similar PCR amplification patterns were obtained with primers related to the other platelet-specific alloantigens (HPA-3 and -5).
Figure 1. Examples of amplification in HPA-1a system. The upper band corresponds to the amplification product of the CRP gene (internal control of 440 bp). The lower band corresponds to the amplification product of HPA-1a gene (189 bp) situated between 123 and 246 bp bands of molecular weight markers.Lane1: negative control; lanes 2, 3, 4, 6, and 7: individuals with HPA-1a allele; lane 5: individual without HPA-1a allele; lane 8: molecular weight marker of 123 bp.
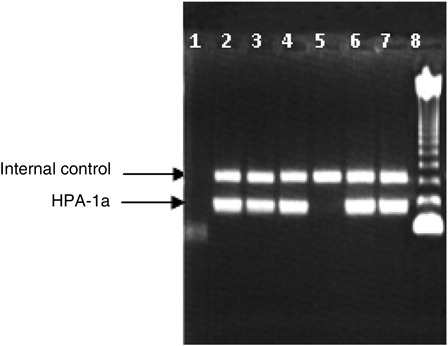
Using this procedure, we typed 212 Southern Tunisian blood donors for HPA-1, -3, and -5 systems. Tables and summarize genotype and allele frequencies. There is no significant deviation from the Hardy–Weinberg equilibrium in any of the studied systems in this population (respectively, χ2 0.55; 0.2 and 0.9).
Table 2. Genotype frequencies of HPA-1, -3, and -5
Table 3. Frequencies of HPA alleles in different ethnic groups
showed also a comparison between our results and those observed in other studied populations.
Discussion
The present study is the first to investigate allele frequency of the HPA-1, -3, and -5 systems in Southern Tunisian population and serves as an outline for future clinical research associated with platelet disorders in this group. A HPA-typed platelet of 212 volunteers and regular blood donor registry was established. In our blood donors, a and b alleles frequencies of the three systems HPA-1, -3, and -5 are similar to those characterizing Caucasian populations.
PCR-SSP, adapted in our laboratory, is the most used technique for HPA genotyping.Citation7 It is a rapid, simple and non-laborious technique. PCR products are visualized directly after gel migration. The required reagents and equipments are standardized, which make PCR-SSP relatively inexpensive.Citation13 In routine, the cost of HPA genotyping may be reduced without compromising the resolution. This is achieved by limiting the testing to the most involved systems in clinical pathology in a given population.Citation14 For this reason, our choice is limited to three systems HPA-1, -3, and -5: the most involved among Caucasians.Citation7 Genotyping results of southern Tunisia population are shown in . The comparison between our results and the literature showed that
- HPA-1a allele frequency varies from north to south in Tunisia. In the centre, HPA-1a frequency was the lowest value observed in the literature (0.59 and 0.66). These frequencies are significantly lower than those found in our study.Citation24,Citation25 When compared to north Tunisia, our HPA-1a frequency showed no significant difference.Citation23 Alleles distribution could be explained by predominant Berber influence in North and South, consanguineous marriages and technical considerations. In fact, in our study, blood donors which are negative for HPA-1a allele, were systematically checked and sometimes proved positive, which was not reported in the methodology of other studies.Citation23,Citation26,Citation27
- Compared to other countries, the prevalence of the HPA-1a allele among southern Tunisian (0.814) was lower than that reported for Asians (Chinese, Japanese, and Pakistani)Citation16,Citation19,Citation29 and Brazilians.Citation20 Nevertheless, the prevalence of this allele in south Tunisia was similar to those reported in other Caucasian populations.Citation15,Citation22
- In southern Tunisia, the HPA-3 polymorphism system shows a similar prevalence in comparison with all other populations so far studied.
- When compared to northern Tunisia, HPA-5a frequency in our blood donors was significantly higher.Citation23 Thus, the high frequency of 5b allele in north Tunisia could be explained by a large introduction of this allele through black slaves. The same conclusion was reported in studies on sickle cell anemia with the import of the Benin gene into Tunisia from black Africa.Citation28
- HPA-5 allele frequencies analysis in southern Tunisia revealed a lower 5a allele frequency to those reported for ChineseCitation16 and Japanese.Citation29 No statistically significant difference was found when compared to Berbers of Morocco, French, and Brazilian Caucasians.Citation15,Citation20,Citation22
These findings could be explained by the nature and the diversity of ethnic groups in Tunisian population. Indeed, Berbers, the original inhabitants of Tunisia and western part of North Africa, have been successively colonized and brewed with Phoenicians, Canaan (territory extended from Syria to Egypt through Lebanon and Palestine), Romans, Byzantines, Andalusians, Jews, Black slaves from trade between Sudan and Europe, and Turks.Citation23
Conclusion
The study of allelic polymorphism of HPA is very important, not only for anthropological and genetic reasons, but also to better predict the risk for allo-immunization for HPAs among distinct ethnic groups. The relative importance of each system HPA in anti-platelet allo-immunization varies among ethnic groups. Based on what has already been reported among Caucasians, our choice was limited to three systems HPA-1, -3, and -5.Citation17 Allele frequencies of the three systems in south Tunisia were similar to those observed in Caucasians populations. The risk of immunization would join therefore that of Caucasians but seems to be less important than Northern Tunisia.
This study allowed us to establish a HPA-typed platelet registry of 212 volunteers and regular blood donor. This registry could help in contacting blood donors of rare and negative phenotypes who are involved in platelets donation. The use of typed platelets is highly beneficial and even indispensable in certain diagnostic situations (research and identification of specific anti-platelet allo-antibodies) and/or therapeutic (NAIT).
Acknowledgments
This research was supported by CRTS (Centre Régional de Transfusion Sanguine de Sfax) and CBS (Centre de Biotechnologie de Sfax), Tunisia.
References
- Santoso S, Kiefel V. Human platelet-specific alloantigens: update. Vox Sang. 1998;74:249–53.
- Metcalfe P, Watkins NA, Ouwehand WH, Kaplan C, Newman P, Kekomaki R, et al. Nomenclature of human platelet antigens. Vox Sang. 2003;85:240–245.
- von dem Borne AE, Décary F. ICSH/ISBT Working Party on platelet serology. Nomenclature of platelet-specific antigens. Vox Sang. 1990;58:176.
- Mueller-Eckhardt C, Santoso S, Kiefel V. Platelet alloantigens-molecular, genetic and clinical aspects. Vox Sang. 1994;67:89–93.
- Santoso S, Santoso S, Kiefel V, Masri R, Mueller-Eckhardt C. Frequency of platelet specific antigens among Indonesians. Transfusion 1993;33:739–41.
- Kim HO, Jin Y, Kickler TS, Blakemore K, Kwon OH, Bray PF. Gene frequencies of the five major human platelet antigens in African American, white, and Korean populations. Transfusion 1995;35:863–7.
- Klüter H, Fehlau K, Panzer S, Kirchner H, Bein G. Rapid typing for human platelet antigen systems-1-2-3 and -5 by PCR amplification with sequence-specific primers. Vox Sang. 1996;71:121–5.
- Chiba AK, Bordin JO, Kuwano ST, Figueiredo MS, Carvalho KI, Vieira-Filho JP, et al. Platelet alloantigen frequencies in Amazon Indians and Brazilian blood donors. Transf Med. 2000;10:207–212.
- Aster RH. New approaches to an old problem. Refractoriness to platelet transfusions. Transfusion 1988;28:95–6.
- Godeau B, Fromont P, Seror T, Duedari N, Bierling P. Platelet alloimmunization after multiple transfusions: a prospective study of 50 patients. Br J Haematol. 1992;81:395–400.
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA form human nucleated cells. Nucleic Acid Res. 1988;16:1215.
- Huret JL, Dessen P, Bernheim A. Atlas of genetics and cytogenetics in oncology and haematology. Nucleic Acids Res. 2001;29(1):303–304.
- Skogen B, Bellissimo DB, Hessner MJ, Santoso S, Aster RH, Newman PJ, et al. Rapid determination of platelet alloantigen genotypes by polymerase chain reaction using allele-specific primers. Transfusion 1994;34:955–60.
- Jones DC, Bunce M, Fuggle SV, Young NT, Marshall SE. Human platelet alloantigens (HPAs): PCR-SSP genotyping of a UK population for 15 HPA alleles. Eur J Immunogenet. 2003;30:415–9.
- Ferrer G, Muñiz-Diaz E, Aluja MP, Arilla M, Martinez C, Nogués R, et al. Analysis of human platelet antigen systems in a Moroccan Berber population. Transfus Med. 2002;12:49–54.
- Feng ML, Liu DZ, Shen W, Wang JL, Guo ZH, Zhang X, et al. Etablissement of an HPA-1-to-16-typed platelet donor registry in China. Transfus. Med. 2006;16:369–374.
- Chen DF, Pastucha LT, Chen HY, Kadar JG, Stangel W. Simultaneous genotyping of human platelet antigens by hot start sequence-specific polymerase chain reaction with DNA polymerase AmpliTaq Gold. Vox Sang 1997;72:192–6.
- http://www.ebi.ac.uk/inc/foot.html.
- Bhatti FA, Uddin M, Ahmed A, Bugert P. Human platelet antigen polymorphisms (HPA-1, -2, -3, -4, -5 and -15) in major ethnic groups of Pakistan. Transfus Med. 2009;20:78–87.
- Castro V, Origa AF, Annichino-Bizzacchi JM, Soares M, Menezes RC, Gonçalves MS, et al. Frequencies of platelet-specific allontigens system 1–5 in three distinct ethnic groups in Brasil. Eur J Immunogenet. 1999;26:355–360.
- Mercier P, Chicheportiche C, Dabonian C, Gamerre M, Reviron D. Platelet antigen HPA-5b (Bra) in the Algerian population. Tissue Antigens 1994;43:58–9.
- Mérieux Y, Dehost M, Bernaud J, Raffin A, Meyer F, Rigal D. Human platelet antigens frequencies of platelet donors in the French population deteminated by polymerase chain reaction with sequence specific primers. Pathol Biol. 1997;45:697–700.
- Mojaat N, Halle L, Proulle V, Hmida S, Ben Hamed L, Boukef K, et al. Gene frequencies of human platelet antigens in the Tunisian population. Tissue Antigens 1999;54:201–204.
- Abboud N, Amin H, Ghazouani L, Ben Haj Khalifa S, Ben Khalafallah A, Aded F, et al. Polymorphisms of human platelet alloantigens HPA-1 and HPA-2 associated with severe coronary artery disease. Cardiovasc Pathol. 2010;19(5):302–307.
- Saidi S, Mahjoub T, Slamia LB, Ammou SB, Al-Subaie AM, Almawi WY. Polymorphisms of the human platelet alloantigens HPA-1, HPA-2, HPA-3, and HPA-4 in ischemic stroke. Am J Hematol. 2008;83:570–573.
- Gorgi Y, Sfar I, Ben Abdallah T, Aouadi H, Abderrahim E, Bardi R, et al. Human Platelet Antigens: HPA-1, -2, -3, -4, and -5 Polymorphisms in Kidney Transplantation. Transplant Proc. 2007;39:2568–70.
- Gorgi Y, Sfar I, Ben Aabdallah T, Aouadi H, Abderrahim E, Bardi R, et al. Human platelet antigens polymorphisms and susceptibility of thrombosis in hemodialysis patients. Hemodial Int. 2008;12:331–5. 58.
- Frikha M, Fakhfakh F, Mseddi S, Gargouri J, Ghali L, Labiadh Z, et al. Study of hemoglobin βS haplotype in the Kebili region of southern Tunisia. Transfus Clin Biol. 1998;5:167–173.
- Legler TJ, Köhler M, Mayr WR, Panzer S, Ohto H, Fischer GF. Genotyping of the human platelet antigen systems 1 through 5 by multiplex polymerase chain reaction and ligation-based typing. Transfusion 1996;36(5):426–31.
- Tanaka S, Ohnoki S, Shibata H, Okubo Y, Yamaguchi H, Shibata Y. Genes frequencies of human platelet antigens on glycoprotein IIIa in Japanese. Transfusion 1996;36:813–7.