Abstract
Background
Despite the excellent efficacy results of imatinib treatment in CML patients, resistance to imatinib has emerged as a significant problem. Genetic variations in genes involved in drug transportation might influence the pharmacokinetic and metabolism of imatinib. The genotype of a patient is increasingly recognized in influencing the response to the treatment.
Aim
To investigate the genotype frequencies of single nucleotide polymorphisms (SNPs) G2677T in CML patients undergoing imatinib treatment to determine whether different genotype pattern of these SNPs have any influence in mediating response to imatinib.
Methods
A total of 96 CML and 90 control samples were analyzed for the human multidrug resistance gene 1 (MDR1) gene polymorphism (G2677T) using polymerase chain reaction-restriction fragment length polymorphism technique.
Results
Genotype distribution revealed a significant lower frequency of TT genotype in CML patients and non-significant difference in the GG, GT genotype frequencies between patients and controls (P = 0.004, 0.138, 0.210, respectively). GG genotype was significantly higher in chronic phase (P = 0.046), while GT genotype was significantly higher in Blastic crisis phase (P = 0.002). There was a significant difference in genotype frequency of G2677T among patients showing response and resistance to imatinib in chronic phase (P = 0.02). TT genotype was associated with complete hematological response (P = 0.01), complete cytogenetic response (P < 0.001), and better molecular response with a significant association (P < 0.001). GT genotype was associated with partial hematological response (P = 0.01) and minor cytogenetic response (P < 0.001). Optimal and suboptimal responses were observed for patients with TT genotype (P = 0.003). Failure of drug response was associated with GT genotype (P = 0.02); however, GG had no association with drug response. Multivariate analysis considered GT genotype as independent risk factor for resistance (P = 0.037), while TT genotype as protective factor against resistance to imatinib (P = 0.008).
Conclusion
Determination of MDR1 polymorphisms (G2677T) might be useful in response prediction to therapy with imatinib in patients with CML.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder that comprises 14% of all leukemias. Imatinib-mysylate, also known as Glivec or Gleevec, is frontline therapy for Philadelphia positive (Ph+) CML.Citation1 It specially targets tyrosine kinase domain of the BCR-ABL fusion protein, and blocks its phosphorylation, which is needed for kinase activation and signal transduction.Citation2 The treatment of CML patients with imatinib has resulted in complete cytogenetic response (CCR) rates of 65–85%.Citation3 Despite of its striking efficacy, however, resistance develops over time in many patients. Resistance is more common in patients who start imatinib in late chronic phase and occurs in approximately 70% of patients treated in myeloid blast crisis and in >90% of those in lymphoid blast crisis.Citation2 Several molecular mechanisms leading to imatinib resistance have been proposed; amplification and overexpression of the BCR/ABL gene, point mutations in the ATP-binding site with kinase reactivation,Citation4 or overexpression of the MDR1 gene.Citation5 The human multidrug resistance gene (MDR1, ABCB1) is located on the long arm of seventh chromosome at q21.1 and consists of 28 exons,Citation6 encodes for P-glycoprotein (P-gp) of 170KD, an ATP-binding cassette transmembrane transporter.Citation7 P-gp is well known to be overexpressed in human tumor cells after cancer chemotherapy where it is associated with multidtug resistance and affect the pharmacokinetics of many drugs that are P-glycoprotein substrates.Citation8 Imatinib is a substrate of P-gp-mediated efflux. The up-regulation of drug transporters (ABCB1) is one of specific causes of imatinib resistance, since it can be effluxed through MDR1 transporters,Citation9 resulting in enhanced clearance of drug from the cell, reduced drug availability and drug resistance.Citation2 MDR1 gene is polymorphic, and more than 1279 single nucleotide polymorphisms (SNPs) have been identified.Citation10 SNPs have the potential to affect the expression and function of the P-gp,Citation11,Citation12 could also influence the efficiency of absorption or elimination of imatinib and could explain at least in part variable responses to this drug.Citation9,Citation13 G2677T, is the most common variants in the coding region of ABCB1.Citation11 The G2677T polymorphism was significantly associated with increased or decreased plasma concentration of P-gp substrates.Citation14,Citation15 Previous reports showed that individuals who had the 2677 TT genotype had lower P-gp messenger RNA expression than those who had 2677 GG genotype.Citation16 On the contrary, some pharmacokinetic studies reported an opposite effect of the 2677T mutant allele, i.e. an increase in transport activity compared with that of 2677G allele,Citation17 whereas Tanabe et al.Citation18 reported a non-significant opposite trend for P-gp expression in placenta in relation to the G2677T polymorphism. These contradictions might be due to the presence of different amino acids at position 893, which might have different effects on different drugs. Identifying influential SNPs may allow to predict the drug disposition, response to imatinib, and may be helpful in dose adjustments of imatinib, which should be effective in case of imatinib resistant individuals.Citation9,Citation19
Patients and methods
Patients
A total of 96 Ph+ CML patients admitted to Mansoura Oncology Center (OCMU) during 2012 were eligible in our study. They were treated regularly with (400–600 mg)imatinib. Sixty-six (68.75%) patients were in chronic phase, 18 (18.75%) in accelerated phase, and 12 (12.5%) patients in blastic crises. They were 54 (56.25%) males and 42 (43.75%) females with mean age of 44.44 ± 12.37 years. In addition, 90 healthy subjects with matched age and sex without history of cancer were randomly selected as a control group. A written informed consent was obtained from all subjects prior to their enrollment in this study.
Chronic-phase CML was defined by the presence of less than 10% blasts, less than 20% basophils, and less than 30% blasts plus promyelocytes in peripheral blood or bone marrow and a platelet count of at least 100 000/cmm, with no extramedullary involvement. Accelerated phase was characterized by a proportion of blasts of 10–30%, or blasts and promyelocytes < 30%, basophils > 20%, platelets >100 × 109/l unrelated to therapy, or chromosomal abnormalities other than the Philadelphia chromosome, or progressive splenomegaly. Blast crisis was defined as the presence of >30% blasts in peripheral blood or bone marrow or as evidence for an extramedullary blast infiltration (except liver, spleen, or lymph nodes).Citation3
The evaluation of the response is based on blood counts and differential (hematologic response, HR), on the examination of marrow cell metaphases (cytogenetic response, CgR) and on a quantitative assessment of BCR-ABL transcripts level (molecular response, MolR).Citation20 HR was defined as normalized peripheral blood cell counts (WBC <10 × 109/l and platelet count <450 × 109/l) without evidence of peripheral blasts, promyelocytes, or myelocytes, and without evidence of extramedullary disease including disappearance of palpable splenomegaly lasting for at least 4 weeks. Cytogenetic responses were categorized as complete (CCyR; 0% Ph+ cells in marrow by conventional cytogenetics or fluorescence in situ hybridization), partial (1–35% Ph+ cells in marrow), minor (36–65% Ph+ cells in marrow), minimal (66–95% Ph+ cells in marrow), and none (>95% Ph+ cells in marrow). A major cytogenetic response was defined as the sum of CCyR and partial cytogenetic response (0–35% Ph+ cells in marrow). Complete molecular response was defined by non-detectable, non-quantifiable BCR/ABL mRNA. Major MolR was defined as ratio of BCR-ABL to ABL (or other housekeeping genes) ≤0.1% on the international scale.Citation20,Citation21
An optimal response to imatinib is defined by complete HR and at least minimal CgR (Ph+ <95%) at 3 months, at least partial CgR (Ph+ <35%) at 6 months, complete CgR at 12 months and major MolR (BCR-ABL: ABL ≤ 0.1%) at 18 months. Failure is defined by incomplete HR at 3 months, no CgR (Ph+ > 95%) at 6 months, less than partial CgR (Ph+ > 35%) at 12 months, less than complete CgR at 18 months and loss of a complete HR or a complete CgR. In any other situation, the response is defined suboptimal.Citation20
Patients were regularly monitored on an outpatient basis; biweekly physical examinations, blood counts, and biochemistry were obtained during the first month of imatinib therapy and then monthly until a cytogenetic response was achieved, and then every 3 months thereafter until a CCR was confirmed, bone marrow evaluation was performed every 3 months.
Methods
Cytogenetic analysis
Pretreatment cytogenetic analyses of bone marrow or peripheral blood for Philadelphia chromosome was done; metaphase chromosomes were banded by G-banding technique and Karyotyped according to the International System for Human Cytogenetic Nomenclature. A minimum of 20 metaphases was required to be examined for a patient to be classified as having normal cytogenetics.Citation22
Real-time quantitative polymerase chain reaction
To assess molecular responses, total RNA was extracted from peripheral or bone marrow blood cells. BCR-ABL and internal control transcript levels were quantified using real-time polymerase chain reaction (PCR) analysis (TaqMan) on an ABI prism 7000 sequence detection system (Applied Biosystems, Foster City, CA, USA). Specific PCR products were amplified and detected using dual-fluorescent non-extendable probes labeled with 6-carboxy-fluorescein (FAM), reporter and 6-carboxytetramethylrhodamine (TAMRA), quencher at 5′-end and 3′-end, respectively. The relative mRNA expression of BCR-ABL transcript was calculated using the comparative cycle threshold (Ct) method.Citation23
Analysis of G2677T polymorphism
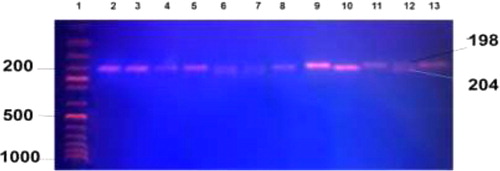
DNA from the peripheral blood of the study subjects were extracted using QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. The G2677T polymorphism was analyzed using a set of primers F5′-TGC AGG CTATAG GTT CCA GG-3′ and R5′-TTT AGTTTG ACT CAC CTT CCC G-3′ to amplify 224-bp fragment. PCRs were performed in a final volume of 25 µl with 0.5 µM of each of the primers (Sigma-Aldrich, St Louis, MO, USA), 400 µM of each dNTPs, 10 mM Tris-HCL, 1.5 mM MgCl2, 50 mM KCl (pH 8.3), and 1 U Taq polymerase (Roche Diagnostics, Mannheim, Germany). PCR amplification consisted of an initial denaturation for 5 minutes at 95°C followed by 40 cycles of denaturation at 95°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 1 minute. The terminal elongation was performed at 72°C for 20 minutes. The amplified PCR products were digestedwith Ban I restriction enzyme (New England, Biolabs) for 1 hour at 37°C. After digestion, the products were electrophoresed on a 2% agarose gel for genotyping. 2677G allele creates site for Ban I enzyme and produces two fragments of 198 and 26 bp whereas 2677T allele was identified by single fragment of 224 bp ().
Statistical analysis
All the statistical analyses were performed with Statistical Package for the Social Sciences (SPSS) 16.0. Mean (±SD) values were used for quantitative data, numbers and percentages were used for qualitative data. Chi-square test was calculated to test the significance of qualitative variables. One-way analysis of variance test were calculated to test for quantitative variables. Multivariate analysis was performed by unconditional logistic regression analysis. The risk associated with a genetic variant was expressed as odds ratio (with a 95% confidence interval). All the P values were two-sided and the level of significance was taken as P < 0.05.
Results
This study comprised 96 CML patients and 90 control subjects. The overall frequencies of the MDR-1 2677 GG, GT, TT, genotypes were 34.4, 46.9, and 18.8%, respectively. There was no significant difference in the GG, GT genotype frequencies, and a significant lower frequency of TT genotype between patients and controls (P = 0.13, 0.21, 0.004, respectively) (). No significant association of G2677T polymorphism was observed with respect to age and sex (P = 0.25, 0.55). When the association of G2677T polymorphisms with clinical phase was considered, GG genotype was significantly higher in chronic phase (27; 40.9%) compared to accelerated/blast crisis (6; 20%) (P = 0.046). Hetrozygous GT genotype was more frequent in blastic crisis (12; 100%) than other phases (P = 0.002). With respect to drug response status, complete hematological response was achieved in 49% of all cases, with significant association with TT genotype (77.8%), while partial hematological response was achieved in 51% of all cases, with significant association with GT genotype (64.4%) (P = 0.010).
Table 1. Distribution of MDR1 G2677T polymorphism in CML patients and control
Cytogenetic response varied significantly between GG, GT, and TT genotypes (P < 0.001). CCR was achieved in 45.8% of all cases, with significantly higher incidence of TT genotype (72.2%), minor cytogenetic response was achieved in 17.7% of all cases with higher incidence of GT genotype (31.1%).
Molecular response was achieved completely in 26%. A better molecular response was observed for patients with TT genotype with a significant association (P < 0.001) ().
Table 2. Patients’ characteristics according to MDR G2677T polymorphisms
Out of 96 CML patients, 50 (52.1%) were responsive (36 in chronic phase and 14 in accelerated/blast crisis) and 46 (47.9%) were resistant (30 in chronic phase and 16 in accelerated/blast crisis) to imatinib with a significant difference in the distribution of G2677T genotypes among sensitive and resistance groups in chronic phase (P = 0.002) (), where TT genotype was associated with optimal and suboptimal response to imatinib (P = 0.003), meanwhile GT was associated with response failure to imatinib (60%; P = 0.026). GG had no association with drug response (P = 0.93) ().
Table 3. Response to Imatinib according to MDR G2677T polymorphisms various in CML phases
Table 4. Response to imatinib according to MDR G2677T polymorphisms
The relationship between G2677T polymorphism and risk of imatinib resistance in CML patients was assessed by means of multivariate analysis that considered GT genotype as independent risk factor for resistance (P = 0.037, odds ratio, OR: 2.519, 95% confidence interval, CI: 1.059–5.990), meanwhile TT genotype as protective factor against resistance to imatinib (P = 0.008, OR: 0.166, 95% CI: 0.044–0.627). However, accelerated/blastic phases were not predictors for imatinib resistance (P = 0.777, OR: 1.139, 95% CI: 0.462–2.811).
Discussion
It is well known that different patients may respond differently to the same drug. The MDR1 polymorphisms might alter P-gp expression and activity toward specific anticancer agents, thereby influencing their therapeutic efficacy; the functional consequences of the changes at positions 2677 is still controversial.Citation24 The SNP G2677T in exon 21 (893 codon) results in the substitution of alanine to serine/threonine in such a manner that the lipophilic residue (Ala) is changed to hydrophilic residue (Ser, Thr) conferring higher resistance to various drugs such as adriamycin and vinblastine.Citation25,Citation26 The frequency of G2677Tgenotypes in this study was similar to that previously reported by Dulucq et al.,Citation19 and dissimilar to Ni et al.,Citation27 and Sailaja et al. Citation2 Interethnic differences in the frequencies of several SNPs have been reported. In particular, G2677T SNPs have been found to differ significantly among different ethnic groups.Citation24 GG genotype was significantly higher in chronic phase. While GT was higher in blast crisis, likewise reported by Sailaja et al.Citation2, who reported that heterozygous 2677GT genotype frequency was increased in blast crisis compared to other phases. GT genotype frequency was associated with partial hematological and minor cytogenetic response, while the frequency of GG genotype was elevated in minimal cytogenetic response in consistent with Sailaja et al.Citation2 TT genotype was associated with CCR. However Ni et al.Citation27 reported a better CCR with GT genotype. A better molecular response was observed for patients with TT genotype with a significant association. Dulucq et al.Citation19 reported a better major molecular response (MMR) for patients with TT genotype. Another study conducted by Kim et al.Citation28, on CML patients, did not find an association between G2677T polymorphism and MMR. We observed a significant difference in the distribution of G2677T genotypes among sensitive and resistance groups in chronic phase likewise reported by Ni et al.Citation27 TT was significantly associated optimal and suboptimal imatinib response that also confirmed by multivariate analysis. However, the study of Gurney et al.Citation9, showed that patients with 2677T homozygotes had higher estimated imatinib clearance compared with other genotypes. Another study on AML patients reported that 2677T allele was associated significantly with shorter relapse time and survival rates compared to heterozygotes.Citation29 In some other studies, haplotype of 2677TT and 3435TT was associated with highest risk of drug resistance in lymphoproliferative diseases.Citation30 These results are surprising in light of impressive report demonstrating an association between decreased P-gp mRNA and 2677T allele.Citation16 Failure of imatinib response was significantly higher with GT genotype that considered as independent risk factor for imatinib resistance by multivariate analysis. This may indicates that 2677 G allele carriers might be at risk for drug resistance. In consistent with our results, Dulucq et al.Citation19 reported that the G allele at 2677 position was significantly associated with worse response in CML patients who were on imatinib therapy. The G allele was reported to have enhanced P-gp expression and was observed to be associated with increased efflux of P-gp substrates. Since imatinib is one of the P-gp substrate, hence G allele was associated with poor response to imatinib in CML patients.Citation2 In line with previous data, studies on G2677T polymorphism have yielded contradictory results, possibly due to small sample sizes and the different ethnic groups.Citation24,Citation31 In conclusion, the results of our study demonstrated a significant association of the SNPs polymorphisms with imatinib efficacy. Hence, genotyping of MDR1 gene polymorphism (G2677T) might be helpful in planning the individualized therapy based on the genotype. Further genotype analysis of other ABCB1 polymorphisms in patients treated with imatinib may be used as a basis for studies on the relationship between ABCB1 genotypes and drug efficacy and may provide some insight into who is likely to respond optimally to imatinib.
References
- Hochhaus A. Chronic myelogenous leukemia (CML) resistance to tyrosine kinase inhibitors. Ann Oncol. 2006;17:274–9.
- Sailaja K, Surekha D, Rao DN, Raghunadharao D, Vishnupriya S. Association of MDR1 gene polymorphism (G2677T) with chronic myeloid leukemia Biology and Medicine. 2010;2 (4):17–21.
- Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, et al. Dasatinib in imatinib resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354(24):2531–41.
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80.
- Mahon FX, Belloc F, Lagarde V, Chollet C, Moreau-Gaudry F, Reiffers J, et al. MDR1 gene overexpression confers resistance to imatinib mesylate in leukemia cell line models. Blood. 2003;101:2368–73.
- Chen CJ, Clark D, Ueda K, Pastan I, Gottesman MM, Roninson IB. Genomic organization of the human multidrug resistance (MDR1) gene and origin of P-glycoproteins. J Biol Chem. 1990;265:506–14.
- Ueda K, Cornwell MM, Gottesman MM, Pastan I, Roninson IB, Ling V, et al. The MDR1 gene, responsible for multidrug resistance, codes for P-glycoprotein. Biochem. Biophys. Res. Commun. 1986;141:956–62.
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58.
- Gurney H, Wong M, Balleine RL, Rivory LP, McLachlan AJ, Hoskins JM, et al. Imatinib deposition and ABCB1(MDR1, Pglycoprotein) genotype. Clin Pharmacol Ther. 2007;82(1):33–40.
- Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta. 2009;1794(5):860–71.
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A ‘‘silent’’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–8.
- Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;97(7):3473–8.
- Hamidovic A, Hahn K, Kolesar J. Clinical significance of ABCB1 genotyping in oncology. J Oncol Pharm Pract. 2010;16:39–44.
- Gervasini G, Pharma D, Carrillo JA, Garcia M, Jose CS, Cabanillas A, et al. Binding Cassette B1 (ABCB1) (Multidrug Resistance 1) G2677t/a Gene polymorphism is associated with high risk of lung cancer. Adenosine Triphosphate. Cancer 2006;107(12):2850–6.
- Siegsmund M, Brinkmann U, Schaffeler E, Weirich G, Schwab M, Eichelbaum M, et al. Association of the Pglycoprotein transporter MDR1 (C3435T) polymorphism with the susceptibility to renal epithelial tumors. J Am Soc Nephrol. 2002;7:1847–54.
- Lamba J, Strom S, Venkataramanan R, Thummel KE, Lin YS, Liu W, et al. MDR1 genotype is associated with hepatic cytochrome P4503A4 basal land induction phenotype. Clin Pharmacol Ther. 2006;79:325–38.
- Kurata Y, Ieiri I, Kimura M, Morita T, Irie S, Urae A, et al. Role of human MDR1 gene polymorphism in bioavailability and interaction of digoxin, a substrate of P-glycoprotein. Clin Pharmacol Therapeut. 2002;72:209–19.
- Tanabe M, Ieiri I, Nagata N, Inoue K, Ito S, Kanamori Y, et al. Expression of P-glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)-1 gene. J Pharmacol Exp Therapeut. 2001; 297(3):1137–43.
- Dulucq S, Bouchet S, Turcq B, Lippert E, Etienne G, Reiffers J, et al. Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood. 2008;112(5):2024–7.
- Baccarani M, Castagnetti F, Gugliotta G, Palandri F, Soverini S; European Leukemia Net. Response definitions and European Leukemianet Management recommendations. Best Pract Res Clin Haematol. 2009;22(3):331–41.
- Druker BJ, Lee SJ. Chronic leukemias. In: Devita VT, Hellman S, Rosenberg SA (eds.) Cancer, principles and practice of oncology. 7th ed. Philadelphia: Pubmed Lippincott; 2005. p. 2124.
- Shaffer LG, Slovak ML, Campbell LJ. ISCN 2009: International System for Human Cytogenetic Nomenclature: Recommendations of the International Standing Committee on Human Cytogenetics Nomenclature. 3rd ed. Basel, Switzerland: Karger Publishing; 2009, pp 364–372.
- Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67:9609–12.
- Kimchi-Sarfaty C, Marple AH, Shinar S, Kimchi AM, Scavo D, Roma MI, et al. Ethnicity-related polymorphisms and haplotypes in the human ABCB1 gene. Pharmacogenomics. 2007;8(1):29–39.
- Kioka N, Tsubota J, Kakehi Y, Komano T, Gottesman MM, Pastan I, et al. P glycoprotein gene (MDR1) cDNA from human adrenal. normal P-glycoprotein carries Gly185 with an altered pattern of multidrug resistance. Biochem Biophys Res Commun. 1989;162:224–31.
- Kim RB, Leake BF, Choo EF, Dresser GK, Kubba SV, Schwarz UI, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70(2):189–99.
- Ni LN, Li JY, Miao KR, Qiao C, Zhang SJ, Qiu HR, et al. Multidrug resistance gene (MDR1) polymorphisms correlate with imatinib response in chronic myeloid leukemia. Med Oncol. 2011;28:265–9.
- Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2009; 15:4750–8.
- Van Den MM, Heuvel-Eibrink EA, Weimer MJ, De Boevere B, Van Der Holt PJ, Vossebeld R, et al. MDR1 gene related clonal selection and P-glycoprotein function and expression in relapsed or refractory acute myeloid leukemia. Blood. 2001;97:3605–11.
- Goreva OB, Grishanova AY, Domnikova NP, Mukhin OV, Lyakhovich VV. MDR1 gene C1236T and C6+139T polymorphisms in the Russian population: associations with predisposition to lymphoproliferative diseases and drug resistance. Bull Exp Biol Med. 2004;138:404–6.
- Tang K, Ngoi SM, Gwee PC, Chua JM, Lee EJ, Chong SS, et al. Distinct haplotype profiles and strong linkage disequilibrium at the MDR1 multidrug transporter gene locus in three ethnic Asian populations. Pharmacogenetics. 2002;12:437–50.