Abstract
A20 is a repressor of NF-κB and was recently shown to be frequently inactivated by deletions or mutations in several types of lymphomas including T-cell lymphoma. Little is known about the characteristics of A20 mutations in T-cell acute lymphoblastic leukemia (T-ALL). In this study, we analyzed A20 polymorphisms and characterized their features in 11 cases with T-ALL, 30 samples from healthy Chinese individuals, and 3 cells lines including CCRF-CEM, Molt-4, and Toledo cells. Two frequent A20 polymorphisms were found: a CCT deletion at position 12384 and a nucleotide exchange (A to C) at position 13751 (rs2307859 and rs661561). The homozygous form (CC) of rs661561 was detected in all 10 cases with detectable T-ALL, while only 80% (24/30) of the healthy controls had this genotype. We found one T-ALL case without the above frequent single-nucleotide polymorphisms (SNPs) in which a T to G mutation at position 12486 was found, which results in an amino acid exchange (Phe127Cys; rs2230926). Similar results were found in Molt-4 cells, which lack the frequent SNPs but have a heterozygous polymorphism at position 13749 (C > T) (rs5029948). Interestingly, the T-ALL case with the Phe127Cys mutation and Molt-4 cells demonstrated a high A20 copy number as measured by real-time polymerase chain reaction amplification with three primer sets that cover different regions of the A20 gene, corresponding to a high A20 and low NF-κB expression level. In conclusion, we characterized the features of A20 polymorphisms in T-ALL, and found that a low frequency A20 mutation, which was thought to be involved in malignant T-ALL development, might function differently in T cell lymphomas.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL), which mainly occurs via the proliferation of malignant T-cell clones, is an aggressive malignancy that does not respond well to chemotherapy and has poorer prognosis than its B-cell counterpart.Citation1,Citation2 T-ALL accounts for 15% of newly diagnosed ALL cases in children and 20–25% of ALL cases in adults.Citation3 Complex acquired genetic aberrations include chromosomal translocations, gene rearrangements and mutations resulting in the abnormal expression of oncogenes such as Notch1, TAL1, and BCL11B, which may be associated with advanced disease and resistance to treatment.Citation4–Citation10 Recently, a number of studies have shown that genetic alterations in A20, a NF-κB negative regulator, are frequently found in lymphomas, suggesting that it may be a linker between the altered immune response and leukemogenesis.Citation11–Citation18
The A20 gene is also known as tumor necrosis factor-α (TNFα)-induced protein 3 (TNFAIP3), which was first discovered in 1990 by Dixit and colleagues as a cytokine-induced gene in human umbilical vein endothelial cells.Citation11,Citation19 The A20 gene is located on chromosome 6q23.3, and its cDNA sequence is 4440 bp long with an open reading frame of 2370 nucleotides that encodes a protein predicted to contain 790 amino acids. A20 contains seven zinc finger (ZnF) domains in its C-terminus including one that functions as an E3 ligase, and an OUT (ovarian tumor) domain is embedded in its N-terminus.Citation18,Citation19
A20 has been reported to be a ubiquitin-editing enzyme with several functions. Although A20 was initially described as an inhibitor of TNF-induced cell death,Citation20,Citation21 subsequent studies demonstrated that A20 overexpression inhibits NF-κB activation in response to different stimuli.Citation22 Increasing data support the notion that A20 is a tumor suppressor. When re-expressed in a lymphoma-derived cell line with no functional A20 alleles, wild type but not mutant A20 resulted in cell growth suppression and apoptosis induction accompanied by the downregulation of NF-κB activation. In A20-deficient cells, the suppression of cell growth and NF-κB activity due to A20 re-expression depended, at least in part, on cell surface-receptor signaling including that of the TNF receptor. These findings indicate that the uncontrolled NF-κB signaling caused by a loss of A20 function is involved in the pathogenesis of B-cell lymphoma subsets, and this loss may contribute to B-cell lymphoma pathogenesis by causing supra-physiological NF-κB activation, which, in turn, has oncogenic properties including inhibiting apoptosis and promoting cell proliferation.Citation14,Citation15
A20 is frequently inactivated by deletions and/or mutations in several lymphoma subtypes including marginal zone lymphoma, diffuse large B-cell lymphoma (DLBCL), follicular lymphoma, extranodal marginal zone lymphoma of mucosa-associated lymphoid tissue, primary mediastinal B-cell lymphoma, Hodgkin's lymphoma and T-cell lymphomas, including Sezary syndrome.Citation12–Citation18 Recently, bi- and monoallelic deletions of A20 in a high proportion of Sezary syndrome patients and biallelic A20 deletion in the Sezary syndrome-derived cell line SeAx was identified. Furthermore, A20 inhibition activates the NF-κB pathway, thereby increasing the proliferation of normal T cells.Citation12 Little is known about the incidence and characteristics of A20 polymorphisms and mutations in T-ALL patients; thus, in this study, we identified four A20 polymorphisms and characterized their features in T-ALL.
Materials and methods
Samples
The samples used in this study were derived from 11 newly diagnosed, untreated patients with T-ALL including nine males and two females (4–55 years old; median age: 32.5 years). Thirty healthy individuals including 18 males and 12 females (22–65 years old; median age: 32 years) served as controls. Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood by Ficoll-Paque gradient centrifugation. RNA and DNA extraction and cDNA synthesis from PBMCs were performed according to the manufacturer's instructions. All human peripheral blood samples were obtained with consent from the human subjects. All procedures were conducted according to the guidelines of the Medical Ethics Committee of the Health Bureau of Guangdong Province in China, and ethical approval was obtained from the Ethics Committee of Medical School of Jinan University for this study.
Polymerase chain reaction and sequencing
To amplify different genomic DNA domains that also cover exons 2–9 (coding region) of the A20 gene according the structure of the A20 gene, nine primer pairs were purchased ().Citation12 Polymerase chain reaction (PCR) was performed as previously described study, and the leukemia T-cell lines CCRF-CEM and Molt-4, the B-cell lymphoma cell line Toledo, and a negative control (non-template) were included in each reaction.Citation5,Citation12 The PCR products were used in the A20 coding sequence mutation analysis by direct sequencing using the BigDye Terminator v3.1 Cycle Sequencing kit (Perkin Elmer, ABI) and the ABI PRISM 3100-Avant genetic analyzer. The sequences from different T-ALL samples, cell lines and healthy individuals were analyzed with BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify polymorphisms or mutations in the A20 gene.
Real-time quantitative PCR
To compare the A20 DNA copy number, we used three pairs of primers that randomly cover different regions (4011–4319, 7170–7477 and 11728–12024) of the A20 gene () to detect the A20 copy number compared with the RAG2 gene copy number (reference gene) by real time PCR. PCR was performed as previously described.Citation5,Citation9,Citation23 In addition, to compare the A20 and NF-κB mRNA expression levels, real-time reverse transcriptase PCR was performed using specific A20 and NF-κB primers (). The relative amount of the genes of interest and β2M reference gene was measured in two independent assays. Specific amplification of the PCR products was analyzed by melting curve analysis. The data are presented as the relative expression of the genes of interest compared to the internal control gene as determined by the 2(−△CT) method.Citation9–Citation10,Citation23
Results and discussion
There are numerous reports of different A20 mutations and deletions in lymphocytic malignancies; however, the frequency of the reported A20 abnormalities is different in different reports, and the most frequent A20 mutations are found in B-cell lymphoma. In addition, A20 mutations were also detected in T-cell lymphomas, including Sezary syndrome.Citation12–Citation18 However, little is known about the A20 molecular alterations in T-ALL.
We used nine primer pairs, which cover the A20 coding region (exon 2 to exon 9), to amplify segments of A20 genomic DNA, and positive PCR products were confirmed by sequencing.Citation12 Recent studies from different countries have reported that A20 is frequently inactivated by deletions and/or mutations in several B- and T-cell lymphoma subtypes, including Sezary syndrome.Citation12–Citation18 However, in this study, we identified four polymorphisms in 11 T-ALL cases, 30 healthy Chinese individuals, and the cell lines CCRF-CEM, Molt-4, and Toledo (DLBCL). The details and frequencies of the polymorphisms are listed in .
Table 1. Details of primers used for PCR and real-time PCR
Table 2. Frequency of A20 gene polymorphisms in T-ALL and healthy individuals
Frequent A20 gene polymorphisms were found including a CCT deletion at position 12384 (located in intron 2 −15 to −18 from exon 2) and an A to C nucleotide exchange position 13751 (located on intron 5) in 10 out of 11 T-ALL cases with both polymorphisms corresponding to a single nucleotide polymorphism (SNP) (IDs: rs2307859 and rs661561, respectively) ( and ).Citation24 Moreover, both SNPs were identified in all of the healthy control samples. These results are similar to the findings of Musone et al. who reported the same SNPs in normal controls and multiple autoimmune diseases.Citation24 Therefore, this result may indicate that there is no significant association between these SNPs and T-ALL. However, the actual difference in the rs661561 SNP between T-ALL patients and healthy controls is that the homozygous form was detected in all 10 cases with T-ALL (100%), and only 80% (24/30) of the healthy controls were homozygous, while the remaining samples were heterozygous at position 13751 (); however, there had no statistical significance of the frequencies in both homozygous or heterozygous polymorphism between T-ALL and control groups (P = 0.125, P = 0.125), this may due to the limit samples, the association between the rs661561 SNP and T-ALL requires further investigation with more numerous samples. There are no reports regarding the distribution of this A20 polymorphism in the Chinese population, and whether there is any influence on the A20 biological function from the heterozygous or homozygous polymorphisms at this or any other position requires further investigation. More interestingly, we found one T-ALL case without the above mentioned frequent SNPs that had a T to G mutation at position 12486 (), which is located in exon 3 and results in an amino acid (at 127 position) exchange from phenylalanine to cysteine (Phe127Cys). This mutation also corresponds to SNP rs2230926, which is the most common coding polymorphism in autoimmune diseases.Citation24 Although it has been reported that this SNP (rs2230926) is detected in healthy individuals in USA.Citation24 However, we identified this mutation in only two healthy individuals in Chinese in this study (2/30) (). There appeared to be a low incidence of rs2230926 in the Chinese cases in this study, although a report demonstrated that the rs5029924, rs5029937, and rs2230926 SNPs in A20 are significantly associated with Chinese systemic lupus erythematosus patients.Citation25 There are few studies describing the incidence of mutations or SNPs in the A20 gene in the Chinese population. Some of the A20 SNPs, such as rs10499194, rs610604, rs7753873, rs5029928, rs9494885, and rs5029939, which are associated with autoimmune diseases in Europeans, were genotyped in Vogt-Koyanagi-Harada (VKH) syndrome, Behcet's disease (BD), Fuchs' heterochromic iridocyclitis and left ventricular hypertrophy (LVH) patients.Citation26–Citation29 Among these SNPs, one strong A20 risk SNP (rs7753873) and two weak A20 risk SNPs (rs10499194 and rs7753873) were identified in BD.Citation26 Moreover, the A20 rs5029939 SNP may be related to a protective genetic marker for the development of LVH in patients with hypertension.Citation27 The frequency of the TT genotype in rs9494885 was markedly lower in VKH disease patients,Citation28 while the five SNPs rs10499194, rs610604, rs7753873, rs5029928, and rs9494885 were not associated with Fuchs' syndrome in a Chinese Han population.Citation29 These results may indicate that the A20 polymorphisms associated with disease susceptibility are different in different ethnic groups. However, further research involving more samples is needed to determine representative results.
Figure 1. The deletion of CCT (rs2307859) at position 12384–12386 in the A20 gene. (A) T-ALL case with a CCT deletion. The arrow indicates the deletion position. (B) T-ALL case with a wild type sequence. The arrow indicates the wild-type sequence with a CCT at position 12384–12386 in the A20 gene.
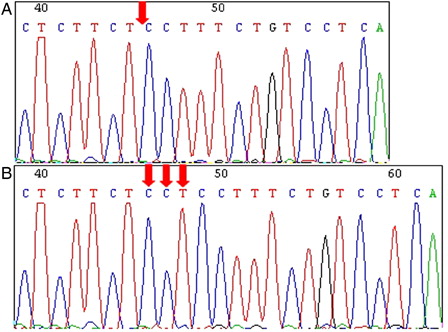
Figure 2. Polymorphism at position 13751 (rs661561) in the A20 gene. (A) T-ALL case with a wild-type sequence. (B) Homozygous A > C nucleotide exchange at position 13751 in a T-ALL case. (C) Heterozygous A > C nucleotide exchange at position 13751 in the Toledo cell line.
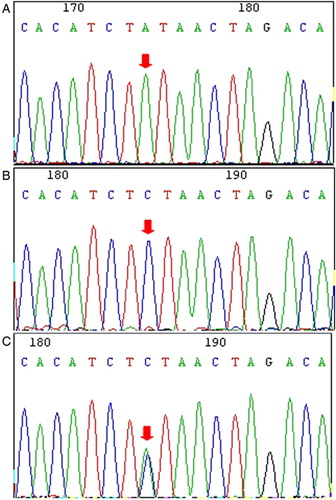
Figure 3. Polymorphism at positions 12486 (rs2230926) and 13749 (rs5029948) in the A20 gene. (A) Wild-type sequence at position 12486 (arrow) in the A20 gene in a T-ALL case. (B) Heterozygous polymorphism (T > G) at position 12486 (arrow) in the A20 gene in a T-ALL case. (C) Wild-type sequence at position 13749 in the A20 gene in a T-ALL case. (D) Heterozygous C > T nucleotide exchange at position 13749 in the A20 gene in Molt-4 cells.
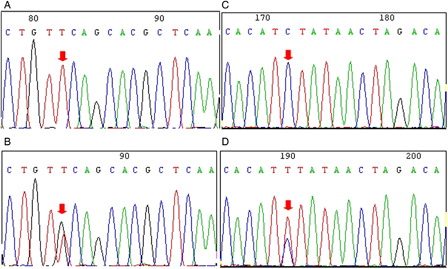
As a control in this study, we also analyzed the A20 polymorphisms in two T-cell lines (CCRF-CEM and Molt-4) and a B-cell line (Toledo). While the same polymorphism characteristics were found in CCRF-CEM and Toledo cells, we found that Molt-4 cells do not contain the frequent SNPs but have a heterozygous polymorphism at position 13749 (C > T) (), which was also identified as a SNP (rs5029948) located in intron 5.Citation24 However, this SNP was not identified in the T-ALL or healthy individuals of the Chinese population in this study, which may be a result of racial diversity.
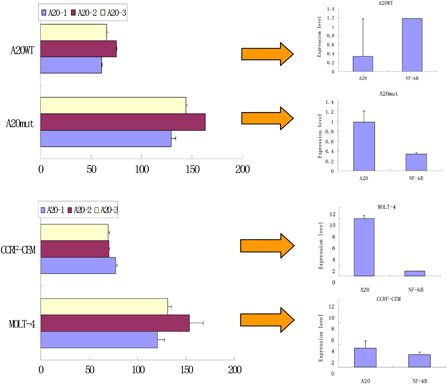
A20 polymorphisms may influence its biological function, e.g. A20 polymorphisms (rs2230926 and rs610604) are associated with the response to TNF blockers in psoriasis,Citation30 and A20 mutations have been suggested to be associated with constitutive NF-κB activation in Sezary syndrome.Citation12,Citation16 It may be interesting to compare the different levels of the expression of A20 and NF-κB in the T-ALL case that lacks the rs2307859 and rs661561 SNPs but has a Phe127Cys A20 mutation. Thus, we compared the A20 DNA copy number and A20 and NF-κB mRNA expression level between two pairs of samples and cell lines including the T-ALL case with the Phe127Cys mutation vs. a T-ALL case without a mutation and the Molt-4 vs. CCRF-CEM cell lines. Interestingly, the T-ALL case with the mutation and the Molt-4 cells (with rs5029948) demonstrated a high A20 DNA copy number as detected by three real-time PCR amplifications using primers that cover different regions of the A20 gene, corresponding to a high A20 and low NF-κB expression level (). This result may imply that such a mutation (rs2230926) or polymorphism (rs5029948) may be related to maintaining an A20 expression level that inhibits NF-κB expression and regulates the immune response in T-ALL.Citation11,Citation14,Citation24,Citation31,Citation32 However, this hypothesis is based only on results from a limited case analysis, and further research involving more samples is needed to determine representative results.
Figure 4. Comparison of the A20 copy number and A20 and NF-κB expression level in leukemia T cells with different polymorphisms in the A20 gene by real-time PCR. A20 WT: a T-ALL case without a Phe127Cys A20 mutation; A20 mut: a case with a Phe127Cys mutation in A20; A20-1: real-time PCR using the A20-4011/4319 primer pair; A20-2: real-time PCR using the A20-7170/7477 primer pair; A20-3: real-time PCR using the A20-11728/12024 primer pair.
In conclusion, we characterized the features of A20 polymorphisms in T-ALL, identified two frequent polymorphisms, and found an A20 mutation in a T-ALL case with an altered A20 and NF-κB expression pattern. The characteristic low frequency A20 mutation in T-ALL is similar to findings in chronic lymphocytic leukemia (CLL),Citation33 where it is thought that malignant CLL development differs from most other B-cell malignancies that show frequent A20 inactivation.Citation34 However, an ongoing study is underway to follow up on the predictive value of this mutation to elucidate the contribution of A20 to the molecular pathogenesis of T-ALL.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Nos. 30871091 and 91129720), the Guangdong Science & Technology Project (2012B050600023), and Science and Technology Innovation Key Project of Guangdong Higher Education Institutes (kjcxzd1013).
References
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–90.
- Morris JC, Waldmann TA, Janik JE. Receptor-directed therapy of T-cell leukemias and Lymphomas. J Immunotoxicol. 2008;5:235–48.
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–48.
- Vanura K, Vrsalovic MM, Le T, Marculescu R, Kusec R, Jäger U, et al. V(D)J targeting mistakes occur at low frequency in acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:725–36.
- Lin C, Zheng H, Wang C, Yang L, Chen S, Li B, et al. Mutations increased overexpression of Notch1 in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 2012;12:13.
- Patel B, Kang Y, Cui K, Litt M, Riberio MS, Deng C, et al. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia 2014;28:349.
- Huang X, Shen Q, Chen S, Chen S, Yang L, Weng J, et al. Gene expression profiles in BCL11B-siRNA treated malignant T cells. J Hematol Oncol. 2011;4:23.
- Huang X, Du X, Li Y. The role of BCL11B in hematological malignancy. Exp Hematol Oncol. 2012;1:22.
- Huang X, Chen S, Shen Q, Chen S, Yang L, Grabarczyk P, et al. Down regulation of BCL11B expression inhibits proliferation and induces apoptosis in malignant T cells by BCL11B-935-siRNA. Hematology. 2011;6:236–42.
- Chen Y, Liu S, Shen Q, Zha X, Zheng H, Yang L, et al. Differential gene expression profiles of PPP2R5C-siRNA-treated malignant T cells. DNA Cell Biol. 2013;32:573–81.
- Zhang F, Yang L, Li Y. The role of A20 in the pathogenesis of lymphocytic malignancy. Cancer Cell Int. 2012;12:44.
- Braun FC, Grabarczyk P, Möbs M, Braun FK, Eberle J, Beyer M, et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sézary syndrome. Leukemia 2011;25:1494–501.
- Chanudet E, Huang Y, Zeng N, Streubel B, Chott A, Raderer M, et al. TNFAIP3 abnormalities in MALT lymphoma with autoimmunity. Br J Haematol. 2011;154:533–5.
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–6.
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–21.
- Honma K, Tsuzuki S, Nakagawa M, Tagawa H, Nakamura S, Morishima Y, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114:2467–75.
- Novak U, Rinaldi A, Kwee I, Nandula SV, Rancoita PM, Compagno M, et al. The NF-κB negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113:4918–21.
- Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–9.
- Dixit VM, Green S, Sarma V, Holzman LB, Wolf FW, O'Rourke K, et al. Tumor necrosis factor-alpha induction of novel gene products in human endothelial cells including a macrophage-specific chemotaxin. J Biol Chem. 1990;265:2973–8.
- Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer 2010;10:332–41.
- Opipari AW Jr., Hu HM, Yabkowitz R, Dixit VM. The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J Biol Chem. 1992;267:12424–7.
- Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci USA. 1996;93:6721–5.
- Li Y, Chen SH, Yang L, Chen S, Lin C, Wang L, et al. Change in expression pattern of TCR-CD3 complex in patients with multiple myeloma. Hematology. 2011;16:143–50.
- Musone SL, Taylor KE, Nititham J, Chu C, Poon A, Liao W, et al. Sequencing of TNFAIP3 and association of variants with multiple autoimmune diseases. Genes Immun. 2011;12:176–82.
- Zhong H, Li XL, Li M, Hao LX, Chen RW, Xiang K, et al. Replicated associations of TNFAIP3, TNIP1 and ETS1 with systemic lupus erythematosus in a southwestern Chinese population. Arthritis Res Ther. 2011;13:R186.
- Li H, Liu Q, Hou S, Du L, Zhou Q, Zhou Y, et al. TNFAIP3 gene polymorphisms confer risk for Behcet's disease in a Chinese Han population. Hum Genet. 2013;132:293–300.
- Xue H, Wang SX, Wang XJ, Xin Y, Wang H, Song XD, et al. Variants of tumor necrosis factor-induced protein 3 gene are associated with left ventricular hypertrophy in hypertensive patients. Chin Med J (Engl). 2011;124:1498–503.
- Li H, Liu Q, Hou S, Du L, Zhou Q, Zhou Y, Kijlstra A, Yang P. TNFAIP3 gene polymorphisms in a Chinese Han population with Vogt-Koyanagi-Harada syndrome. PLoS One. 2013;8:e59515.
- Li H, Hou S, Du L, Zhou Q, Kijlstra A, Liu Q. Polymorphisms of TNFAIP3 Gene in a Chinese Han Population with Fuchs Heterochromic Iridocyclitis. Ophthalmic Genet. 2013 Mar 6. [Epub ahead of print].
- Tejasvi T, Stuart PE, Chandran V, Voorhees JJ, Gladman DD, Rahman P, et al. TNFAIP3 gene polymorphisms are associated with response to TNF blockade in psoriasis. J Invest Dermatol. 2012;132(3 Pt 1):593–600.
- Song XT, Evel-Kabler K, Shen L, Rollins L, Huang XF, Chen SY. A20 is an antigen presentation attenuator, and its inhibition overcomes regulatory T cell-mediated suppression. Nat Med. 2008;14:258–65.
- Shi L, Chen S, Lu Y, Wang X, Xu L, Zhang F, et al. Changes in the MALT1-A20-NF-κB expression pattern may be related to T cell dysfunction in AML. Cancer Cell Int. 2013;13:37.
- Philipp C, Edelmann J, Bühler A, Winkler D, Stilgenbauer S, Küppers R. Mutation analysis of the TNFAIP3 (A20) tumor suppressor gene in CLL. Int J Cancer. 2011;28:1747–50.
- Frenzel LP, Claus R, Plume N, Schwamb J, Konermann C, Pallasch CP, et al. Sustained NF-kappaB activity in chronic lymphocytic leukemia is independent of genetic and epigenetic alterations in the TNFAIP3 (A20) locus. Int J Cancer. 2011;128:2495–500.
