Abstract
Objectives
The effects of NOD2 single nucleotide polymorphisms (SNPs) on Grade III–IV acute graft-versus-host disease (aGVHD) risk are somewhat contradictory in different studies. The aim of the meta-analysis was to clarify the effects of NOD2 SNPs on the incidence of Grade III–IV aGVHD.
Methods
We searched PubMed, EMBASE, Web of SCIENCE, WanFang and Chinese National Knowledge Infrastructure (CNKI) databases to collect eligible publications. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the association between NOD2 polymorphisms and Grade III–IV aGVHD risk.
Results
A total of nine studies from eight publications met the inclusion criteria and were included in this meta-analysis. Patient NOD2 SNPs were not associated with aGVHD risk. A tendency of higher risk to develop Grade III–IV aGVHD was found in patients with pairs NOD2 SNPs. Subgroup analyses showed that pairs NOD2 SNPs were associated with Grade III–IV aGVHD in the Caucasian population and in identical sibling donors (IS), but not in matched unrelated donors (MUD). In patients who received hematopoietic stem cell transplantation (HSCT) with T-cell depletion and gut decontamination, there was still an association between pairs NOD2 SNPs and Grade III–IV aGVHD risk.
Conclusions
Our meta-analysis suggests that pairs NOD2 SNPs, not patient NOD2 SNPs, may be associated with Grade III–IV aGVHD risk, especially in the Caucasian population. It is also indicated that in pairs NOD2 polymorphisms group, patients who receive HSCT from IS may experience higher risk of Grade III–IV aGVHD.
Introduction
Hematopoietic stem cell transplantation (HSCT) is currently the primary curative treatment for patients with severe hematopoietic diseases. Despite advances have been made by HLA matching, recipients remain susceptible to fatal post-transplantation complications, including disease relapse and the development of GVHD.Citation1 Thus, apart from the HLA genes, scientific interest has been switched to novel genetic factors, which may be related to the occurrence of GVHD.Citation2
Holler et al.Citation3 in 2004 identified SNPs in the NOD2 gene as a predictor for the occurrence of aGVHD. Being a member of proteins involved in intracellular pathogen recognition,Citation4 the NOD2 protein has been described as an intracellular sensor to muramyl dipeptide (a compound of the bacterial cell wall) mediating consecutive nuclear factor-κB activation,Citation5 which is restrictedly expressed in intestinal epithelial cells and cells of monocyte/macrophage lineage.Citation4,Citation6 There are plenty of SNPs in the NOD2 gene, such as SNP8 (R702 W, rs2066844), SNP12 (G908R, rs2066845), and SNP13 (1007fs, rs2066847), leading to gene function alteration.Citation7 Failure of NOD2 function may lead to inefficient clearance of bacterial pathogens, followed by subsequent hyperinflammation, which in turn promotes the development of aGVHD.Citation8 Holler et al.Citation3 also suggested that there was an increased release of cytokines for recipients with the polymorphisms, which led to the activation of recipient dendritic cells and donor T cells. All the above studies suggest that NOD2 variants may be associated with the occurrence of aGVHD.
Several studies have evaluated the associations between NOD2 SNPs and aGVHD risk.Citation8–Citation17 However, the results are somewhat contradictory: some groups suggested that NOD2 gene variants were associated with the incidence of aGVHD,Citation9,Citation11,Citation12,Citation17 whereas other studies did not find such a correlation.Citation8,Citation10,Citation13,Citation15 The current divergence in reported studies may be a result of small sample size, different ethnic backgrounds and unadjusted study design confounders. Therefore, a quantitative synthesis for combing the confusing results from several studies is needed to clarify the association between NOD2 variants and Grade III–IV aGVHD risk after HSCT. In this article, we conducted a meta-analysis of all the available studies to clarify the effects of NOD2 polymorphisms on Grade III–IV aGVHD risk.
Materials and methods
Selection of studies
This meta-analysis was conducted in accordance with the PRISMA guidelines.Citation18 We conducted a computerized literature search from PubMed, EMBASE, Web of SCIENCE, WanFang and Chinese National Knowledge Infrastructure (CNKI) databases by May 30, 2014 using the following terms: ‘hematopoietic stem cell transplantation’ or ‘bone marrow transplantation’ or ‘stem cell transplantation’ in combination with ‘NOD2’ or ‘Nucleotide binding oligomerization domain containing 2’. There was no language limitation. We also screened all eligible studies and their references for other relevant studies. If studies involved overlapping subject groups, only the most complete and largest research was included in this analysis. If there was more than one geographic group in one article, each group was considered as a separate study.
Selection criteria
Studies were eligible for inclusion if they met all of the following criteria: (1) patients had received HSCT for hematological diseases; (2) evaluation of the NOD2 SNPs and Grade III–IV aGVHD risk; (3) data were sufficient to calculate the odds ratio (OR) and the corresponding 95% confidence interval (CI). Reviews, case reports, letters, and any conference abstracts were excluded.
Data extraction
To avoid bias in the data abstraction process, data were independently extracted by two authors (H.Z. and M.J.) using the same data collection form. All data were checked for internal consistency and disagreements were resolved by discussion. Data extracted from these studies included: first author, publication year, study country, ethnicity, number of subjects, underlying disease, age range, stem cell source, donor type, conditioning regimen, GVHD prophylaxis, T-cell depleted, decontamination, SNPs studied, genotyping method, number of cases (who developed aGVHD) or controls (who did not develop aGVHD), and genotype distribution in both groups.
Statistical methods
To quantify the NOD2 variant effects on Grade III–IV aGVHD after HSCT, pooled ORs and their corresponding 95% CIs were calculated in two groups: pairs SNPs (pairs SNPs denoting SNP present in donor and/or recipient) and patient SNPs (patient SNPs denoting SNP present in recipients only). Subgroup analyses were performed to investigate the effects of NOD2 SNPs on Grade III–IV aGVHD risk based on ethnicity and donor type. Moreover, impacts of NOD2 SNPs on Grade III–IV aGVHD risk in studies with T-cell depletion and gut decontamination during HSCT were also analyzed (T-cell depletion in the study of Holler et al.Citation12 was only conducted in a minority of patients, so the two studies were excluded in this analysis).
The degree of heterogeneity between studies was assessed with Chi square-based Q statistic test.Citation19 When the heterogeneity was high (a P < 0.1), a random-effects model was used to pool the OR, otherwise a fixed-effects model was used. Publication bias was conducted using Begg's funnel plots and Egger's test.Citation20,Citation21 All the statistical calculations for this meta-analysis were performed with Stata Statistical Software Version 12.0 (StataCorp LP, College Station, TX, USA), and a P < 0.05 was considered to be statistically significant.
Results
Characteristics of included studies
According to PRISMA,Citation18 a study selection flow chart is shown in . Briefly, a total of 276 articles were identified after initial search, of which 64 were duplicates. After reviewing the abstracts of the remaining 212 articles, 20 publications were further fully read. Finally, nine studies from eight articles (No. 1–9 in )Citation8,Citation9,Citation11–Citation15,Citation17 met the criteria and were included in this meta-analysis, with seven studies (No. 1–3, 5–7, 9) investigating pairs NOD2 SNPs and three (No. 4, 7, 8) investigating NOD2 SNPs of patients only. Among these, there were seven articles for NOD2 SNP8, SNP12, and SNP13 (No. 1–7, 9), and one article investigated rs1078327 (R471C) (No. 8). Of the nine available studies, seven came from Caucasian populations (No. 1–7), one was from an Asian population (No. 8), and one was performed in a mixed population (No. 9). Holler et al. performed an analysis investigating the NOD2 SNPs and Grade III–IV aGVHD risk in 2006 from two geographic cohorts (No. 2 and 3), which we treated as two independent studies. It is worthwhile to point out that in one study (No. 1), whose donors were from both IS and MUD, genotype distribution of IS was insufficient, but that of MUD was sufficient. Therefore, we only included data regarding MUD, and excluded data about IS. The main characteristics of the patients in each study were summarized and showed in .
Table 1. Summary of studies that have examined the association between NOD2/CARD15 SNPs and aGVHD
Table
The association between patient NOD2 SNPs and Grade III–IV aGVHD after HSCT
There were 3 studies (No. 4, 7, 8) including 849 pairs which assessed the effect of patient NOD2 SNPs on Grade III–IV aGVHD risk after HSCT. The ICitation2 value for the studies was 45.2%, with a P value of 0.161. Thus, we chose a fixed-effects model for the above meta-analysis. There was no significant association between patient NOD2 SNPs and Grade III–IV aGVHD after HSCT: OR, 1.02; 95% CI, 0.51–1.52 (). No significant publication bias was found based on both Egger's test and the Begg's funnel plot test (data not shown). As there were no sufficient data about donor type and ethnicity, we did not perform stratified analyses based on donor type and ethnicity.
Table 2. The results of this meta-analysis
The association between pairs NOD2 SNPs and Grade III–IV aGVHD after HSCT
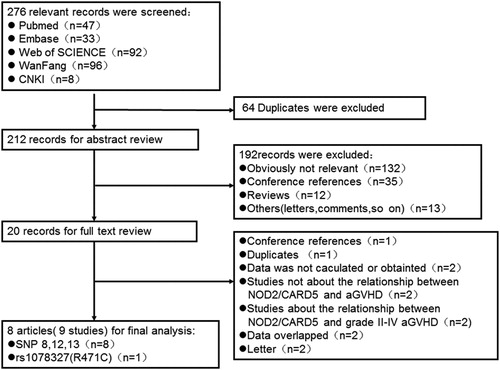
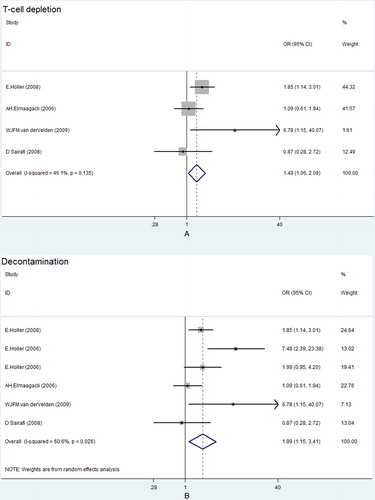
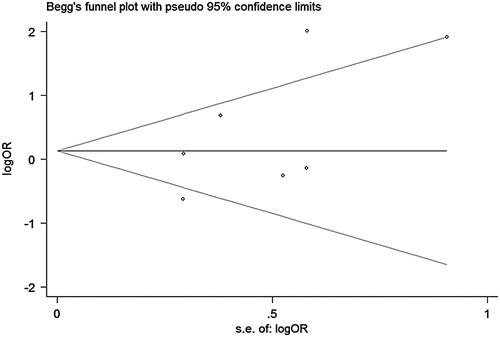
There were 7 studies (No. 1–3, 5–7, 9) including 1721 pairs which assessed the effect of pairs NOD2 SNPs on Grade III–IV aGVHD risk after HSCT. Heterogeneity for the studies was identified (ICitation2, 70.6%; P = 0.002), so we chose a random-effects model for this meta-analysis. Although a statistically significant level was not reached, a tendency of higher risk to develop Grade III–IV aGVHD after HSCT was found for patients who carried any mutations in HSCT pairs (OR, 1.68; 95% CI, 0.98–2.88) (, A). No significant publication bias was found based on both Egger's test (P = 0.506) () and the Begg's funnel plot test (P = 0.230) ().
Figure 2. Forest plot for risk of Grade III–IV aGVHD associated with pairs NOD2 SNPs in all studies (A) and in the Caucasian population (B). OR, odds ratio; CI, confidence interval.
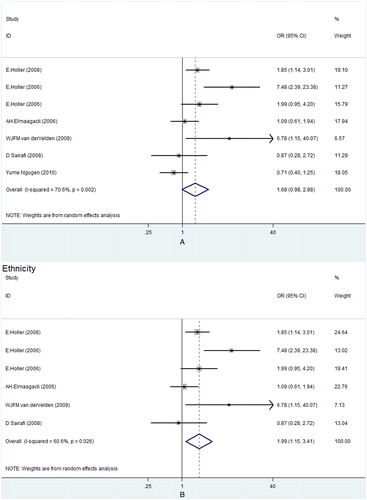
For pairs NOD2 SNPs, subgroup analyses based on ethnicity and donor type were performed. When stratified according to the ethnicity, patients of Caucasian population with pairs SNPs had higher risk to develop Grade III–IV aGVHD (OR, 1.99; 95% CI, 1.15–3.41) (, B). The result of subgroup analysis based on donor type also showed a significant association between SNPs in HSCT pairs and a higher risk of Grade III–IV aGVHD in IS, with an OR of 4.00 (95% CI, 1.50–10.69) (, ). However, no such relationship was found in MUD, with an OR of 1.16 (95% CI, 0.45–2.98) (, ).
Figure 3. Subgroup analysis of pairs NOD2 SNPs and Grade III–IV aGVHD risk in IS (A) and MUD (B). OR, odds ratio; CI, confidence interval.
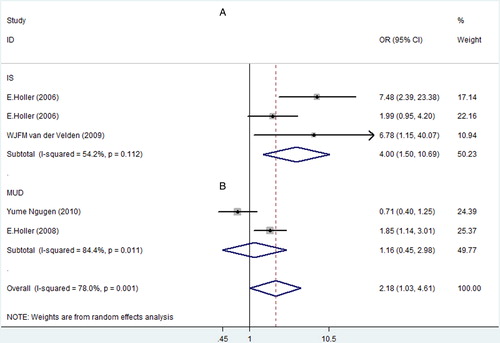
Moreover, pairs NOD2 SNPs had a great correlation with Grade III–IV aGVHD risk in patients with T-cell depletion (OR, 1.49; 95% CI, 1.06–2.09) (, A) and gut decontamination (OR, 1.99; 95% CI, 1.15–3.41) (, B).
Discussion
The incidence of aGVHD is 20–40% of recipients in HLA matched sibling donor grafts, suggesting that factors other than HLA matching play an important role in the initiation of aGVHD.Citation22 Recently, several reports suggest that mutations of the NOD2 gene are associated with an increased incidence of inflammatory bowel disease (IBD).Citation23,Citation24 Based on the fact that the gastrointestinal GVHD and IBD may have some features in common, a number of reports assess the influence of the NOD2 gene mutations on aGVHD risk in patients who receive HSCT. In this article, we comprehensively reviewed the literature on Grade III–IV aGVHD risk and NOD2 SNPs and performed a meta-analysis, for the purpose of identifying patients at a higher risk for aGVHD. To our knowledge, this is the first meta-analysis regarding the association between NOD2 SNPs and Grade III–IV aGVHD.
Figure 5. Funnel plot with pseudo 95% confidence limits about pairs NOD2 SNPs and Grade III–IV aGVHD.
Overall, we found the tendency of higher risk to develop Grade III–IV aGVHD after HSCT for patients in the pairs NOD2 SNPs group. Cytostatic chemotherapy can cause mucosal barrier injury, leading to an uncontrolled inflammatory response and subsequent microbial translocation, which are thought to be important in the initiation of aGVHD.Citation25–Citation27 Altered function due to NOD2 polymorphisms can result in uncontrolled inflammation of the gut mucosaCitation24 and bacterial translocation.Citation17 Moreover, studies of Nau et al.Citation28 and Heinzelmann et al.Citation29 show that NOD2 generates signals for the adaptive immune response, which induces secretion of interleukin-6 and interleukin-12, leading to the differentiation of naïve T cells into effector T cells. These discoveries can explain the strong association of NOD2 SNPs with the incidence of aGVHD. When stratified by ethnicity, the pairs NOD2 polymorphisms had been found to be associated with a higher risk of Grade III–IV aGVHD in the Caucasian population, suggesting a potential role of ethnic differences in genetic background.Citation15 When stratified by donor type, there was a significant association between SNPs in HSCT pairs and a higher risk of Grade III–IV aGVHD in IS, but not in MUD. In the IS transplants, the group in which both donor and recipients revealed identical SNPs will result in an increased gene dose of NOD2 variants.Citation11 What is more, there may be other SNPs which are inherited in a similar way to NOD2 SNPs and therefore favor the associations only in IS transplants.Citation11 Since there were only two studies on MUD, the difference might be caused by sample size, which was too small to have sufficient power to detect the slight effect.
Unfortunately, our results failed to detect the correlation between patient SNPs and Grade III–IV aGVHD risk. There are several explanations for this result. Firstly, Holler et al.Citation12 suggest that combined donor/recipient variants may have the strongest impact on GVHD because of the additive effects of damage in affected cells: both recipient epithelium and donor monocytes represent an additive effect of mutation dose in macrophage-derived cells of the donor and the recipient. Secondly, Tanabe et al.Citation15 conducted a survey in a Japanese population and focused on SNP rs1078327: R471C, which is different from other studies. Recent results have showed that Japanese cohorts are at significantly lower risk for aGVHD than other cohorts,Citation30 and the common NOD2 variants associated with Crohn's disease in Caucasians are different from those in Japanese.Citation31,Citation32 However, when we excluded this study and only analyzed studies of Wermke et al.Citation8 and Elmaagacli et al.,Citation9 the correlation between patient SNPs and Grade III–IV aGVHD risk was still not found (data not shown).
T-cell depletion and gut decontamination are known to be the factors that can greatly influence the incidence of aGVHD after SCT.Citation9,Citation12,Citation14,Citation33 In our meta-analysis, studies of patients with T-cell depletion and gut decontamination during HSCT were analyzed and the results showed that there was still an association between pairs NOD2 SNPs and Grade III–IV aGVHD risk. Martinez et al.Citation34 suggest that adaptive immune reconstitution in T-cell depletion patients is slower than the unmanipulated allogeneic HSCT and may have a higher risk for infection, which may result in higher risk for aGVHD. One article suggests that the use of Campath in T-cell depletion may influence the incidence of aGVHDCitation35 and thereby affect the effect of NOD2 SNPs. Two studies in our study of patients with T-cell depletion partly used Campath.Citation9,Citation11 As we could not get the exact data, the effect of NOD2 SNPs on Grade III–IV aGVHD in T-cell depletion patients should be interpreted with caution.
We also investigated the association between NOD2 SNPs and Grade II–IV aGVHD risk. However, the results could not detect the association between NOD2 SNPs and Grade II–IV aGVHD risk (data not shown), either in pairs SNPs groupCitation9,Citation14,Citation17 or in patient SNPs group.Citation9,Citation10,Citation16 There are several explanations for these results. Firstly, the number of studies about the relationship between Grade II–IV aGVHD and NOD2 SNPs in each group is few, which may restrict the power of these analyses. Secondly, Gruhn et al.Citation10 conducted a survey in children, in which underlying diseases and clinical protocols may be different from other studies. Therefore, the effects of NOD2 SNPs on Grade II–IV aGVHD risk may be more or less masked.
Several limitations in the current meta-analysis exist. Firstly, we did not include unpublished articles to obtain data for analysis, and as a result, publication bias might exist. Secondly, there are only three studies which examined the relationship between patient SNPs and Grade III–IV aGVHD risk. Moreover, some studies were excluded as we could not get sufficient data to calculate the OR,Citation7,Citation36 which may have an impact on our meta-analysis results. The small sample size with decreased power may lead to some of the results being not so robust. In addition, only a few studies investigated the association between donor NOD2 SNPs and risk of Grade III–IV aGVHD. As a result, we could not elaborate the correlation between NOD2 SNPs and Grade III–IV aGVHD risk from the donor side. Thirdly, the between-study heterogeneity in some groups was great, which might be a result of the dissimilarity from different research centers, such as donor type, underlying diseases, HLA matching status, conditioning regimen, and GVHD prophylaxis. In these studies, we applied random-effects models. Therefore, the effect of NOD2 SNPs on Grade III–IV aGVHD should be interpreted with caution.
Although these limitations need to be borne in mind, our meta-analysis suggests that pairs NOD2 SNPs may be associated with Grade III–IV aGVHD risk after HSCT, especially in the Caucasian population. It also indicates that in pairs NOD2 SNPs group, patients who receive HSCT from IS may experience a higher risk of aGVHD. In patients who receive HSCT with T-cell depletion and gut decontamination, there is still an association between pairs NOD2 SNPs and Grade III–IV aGVHD risk. Patient NOD2 SNPs are not associated with Grade III–IV aGVHD risk. Based on the current findings, assessing NOD2 polymorphisms may be clinically useful to estimate Grade III–IV aGVHD risk and further large-scale cohort studies are needed to validate our results.
Disclaimer statements
Contributors HZ: conceiving and designing the study, collecting the data, writing the article in whole or in part; MJ: collecting the data, analyzing the data, writing the article in whole or in part; ZW: collecting the data, interpreting the data; YC: interpreting the data; ZL: interpreting the data; XX: revising the article; SY: revising the article; YT: obtaining funding and/or ethics approval, revising the article.
Funding This study was supported in part by grants from the National Natural Science Foundation of China (No: 30971283, No: 81170502), the Zhejiang Provincial Natural Science Foundation of China (No: LZ12H08001), and Zhejiang Provincial Fund of Health Bureau (No. 2007B122).
Conflicts of interest None.
Ethics approval Ethical approval was not required.
References
- Shaw BE, Madrigal JA, Potter M. Improving the outcome of unrelated donor stem cell transplantation by molecular matching. Blood Rev. 2001;15(4):167–74.
- Sivula J, Turpeinen H, Volin L, Partanen J. Association of IL-10 and IL-10Rbeta gene polymorphisms with graft-versus-host disease after haematopoietic stem cell transplantation from an HLA-identical sibling donor. BMC Immunol. 2009;10:24.
- Holler E, Rogler G, Herfarth H, Brenmoehl J, Wild PJ, Hahn J, et al. Both donor and recipient NOD2/CARD15 mutations associate with transplant-related mortality and GvHD following allogeneic stem cell transplantation. Blood 2004;104(3):889–94.
- Inohara N, Ogura Y, Nunez G. Nods: a family of cytosolic proteins that regulate the host response to pathogens. Curr Opin Microbiol. 2002;5(1):76–80.
- Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278(8):5509–12.
- Berrebi D, Maudinas R, Hugot JP, Chamaillard M, Chareyre F, De Lagausie P et al. Card15 gene overexpression in mononuclear and epithelial cells of the inflamed Crohn's disease colon. Gut 2003;52(6):840–6.
- Granell M, Urbano-Ispizua A, Arostegui JI, Fernandez-Aviles F, Martinez C, Rovira M et al. Effect of NOD2/CARD15 variants in T-cell depleted allogeneic stem cell transplantation. Haematologica 2006;91(10):1372–6.
- Wermke M, Maiwald S, Schmelz R, Thiede C, Schetelig J, Ehninger G et al. Genetic variations of interleukin-23R (1143A>G) and BPI (A645G), but not of NOD2, are associated with acute graft-versus-host disease after allogeneic transplantation. Biol Blood Marrow Transplant. 2010;16(12):1718–27.
- Elmaagacli AH, Koldehoff M, Hindahl H, Steckel NK, Trenschel R, Peceny R et al. Mutations in innate immune system NOD2/CARD 15 and TLR-4 (Thr399Ile) genes influence the risk for severe acute graft-versus-host disease in patients who underwent an allogeneic transplantation. Transplantation 2006;81(2):247–54.
- Gruhn B, Intek J, Pfaffendorf N, Zell R, Corbacioglu S, Zintl F et al. Polymorphism of interleukin-23 receptor gene but not of NOD2/CARD15 is associated with graft-versus-host disease after hematopoietic stem cell transplantation in children. Biol Blood Marrow Transplant. 2009;15(12):1571–7.
- Holler E, Rogler G, Brenmoehl J, Hahn J, Greinix H, Dickinson AM et al. The role of genetic variants of NOD2/CARD15, a receptor of the innate immune system, in GvHD and complications following related and unrelated donor haematopoietic stem cell transplantation. Int J Immunogenet. 2008;35(4–5):381–4.
- Holler E, Rogler G, Brenmoehl J, Hahn J, Herfarth H, Greinix H et al. Prognostic significance of NOD2/CARD15 variants in HLA-identical sibling hematopoietic stem cell transplantation: effect on long-term outcome is confirmed in 2 independent cohorts and may be modulated by the type of gastrointestinal decontamination. Blood 2006;107(10):4189–93.
- Nguyen Y, Al-Lehibi A, Gorbe E, Li E, Haagenson M, Wang T et al. Insufficient evidence for association of NOD2/CARD15 or other inflammatory bowel disease-associated markers on GVHD incidence or other adverse outcomes in T-replete, unrelated donor transplantation. Blood 2010;115(17):3625–31.
- Sairafi D, Uzunel M, Remberger M, Ringden O, Mattsson J. No impact of NOD2/CARD15 on outcome after SCT. Bone Marrow Transplant. 2008;41(11):961–4.
- Tanabe T, Yamaguchi N, Matsuda K, Yamazaki K, Takahashi S, Tojo A et al. Association analysis of the NOD2 gene with susceptibility to graft-versus-host disease in a Japanese population. Int J Hematol. 2011;93(6):771–8.
- van der Straaten HM, Paquay MM, Tilanus MG, van Geloven N, Verdonck LF, Huisman C. NOD2/CARD15 variants are not a risk factor for clinical outcome after nonmyeloablative allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2011;17(8):1231–6.
- van der Velden WJ, Blijlevens NM, Maas FM, Schaap NP, Jansen JH, van der Reijden BA, et al. NOD2 polymorphisms predict severe acute graft-versus-host and treatment-related mortality in T-cell-depleted haematopoietic stem cell transplantation. Bone Marrow Transplant. 2009;44(4):243–8.
- Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62(10):e1–34.
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327(7414):557–60.
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50(4):1088–101.
- Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315(7109):629–34.
- Mullighan C, Heatley S, Doherty K, Szabo F, Grigg A, Hughes T et al. Non-HLA immunogenetic polymorphisms and the risk of complications after allogeneic hemopoietic stem-cell transplantation. Transplantation 2004;77(4):587–96.
- Hampe J, Grebe J, Nikolaus S, Solberg C, Croucher PJ, Mascheretti S et al. Association of NOD2 (CARD 15) genotype with clinical course of Crohn's disease: a cohort study. Lancet 2002;359(9318):1661–5.
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 2001;411(6837):599–603.
- Blijlevens NM. Implications of treatment-induced mucosal barrier injury. Curr Opin Oncol. 2005;17(6):605–10.
- Ferrara JL, Levy R, Chao NJ. Pathophysiologic mechanisms of acute graft-vs.-host disease. Biol Blood Marrow Transplant. 1999;5(6):347–56.
- Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood 2000;95(9):2754–9.
- Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA 2002;99(3):1503–8.
- Heinzelmann M, Polk HJ, Chernobelsky A, Stites TP, Gordon LE. Endotoxin and muramyl dipeptide modulate surface receptor expression on human mononuclear cells. Immunopharmacology 2000;48(2):117–28.
- Oh H, Loberiza FJ, Zhang MJ, Ringden O, Akiyama H, Asai T et al. Comparison of graft-versus-host-disease and survival after HLA-identical sibling bone marrow transplantation in ethnic populations. Blood 2005;105(4):1408–16.
- Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y et al. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology 2002;123(1):86–91.
- Yamazaki K, Takazoe M, Tanaka T, Kazumori T, Nakamura Y. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn's disease. J Hum Genet. 2002;47(9):469–72.
- Beelen DW, Elmaagacli A, Muller KD, Hirche H, Schaefer UW. Influence of intestinal bacterial decontamination using metronidazole and ciprofloxacin or ciprofloxacin alone on the development of acute graft-versus-host disease after marrow transplantation in patients with hematologic malignancies: final results and long-term follow-up of an open-label prospective randomized trial. Blood 1999;93(10):3267–75.
- Martinez C, Urbano-Ispizua A, Rozman C, Marin P, Rovira M, Sierra J et al. Immune reconstitution following allogeneic peripheral blood progenitor cell transplantation: comparison of recipients of positive CD34+ selected grafts with recipients of unmanipulated grafts. Exp Hematol. 1999;27(3):561–8.
- Holler E, Hahn J, Andreesen R, Rogler G, Brenmoehl J, Greinix H, et al. NOD2/CARD15 polymorphisms in allogeneic stem-cell transplantation from unrelated donors: T depletion matters. J Clin Oncol. 2008;26(2):338–9; author reply 9.
- Mayor NP, Shaw BE, Hughes DA, Maldonado-Torres H, Madrigal JA, Keshav S et al. Single nucleotide polymorphisms in the NOD2/CARD15 gene are associated with an increased risk of relapse and death for patients with acute leukemia after hematopoietic stem-cell transplantation with unrelated donors. J Clin Oncol. 2007;25(27):4262–9.