Abstract
Objectives
Despite numerous studies in order to determine the allele frequency and clinical impact of DNA methyltransferase 3 A (DNMT3A) gene mutations in acute myeloid leukemia (AML), reports about the expression analysis of this gene are rare and between the available, differences are evident.
Methods
In this study, we decided to investigate DNMT3A possible expression changes with regard to their mutation and cytogenetic status in a series of 96 AML patients.
Results
Mutations were founded in 17 of the 96 patients (17.7%) and associated with higher age and white blood cell count (P < 0.001). Our mutants have had shorter overall survival (OS) (P < 0.001) and relapse-free survival (RFS) (P = 0.011) than those without. Multivariate analysis showed that DNMT3A mutation is an independent prognostic indicator for OS and RFS (P < 0.001). In relation to expression results, we had over and under expression for our favorable and unfavorable cytogenetic subgroups, respectively (P = 0.005 and P < 0.001, respectively). In intermediate subgroup, total DNMT3A expression did not alter (P = 0.575). Interestingly, we noticed similar expression results for DNMT3A transcript 2, to that of the total.
Discussion and conclusion
In relation to DNMT3A expression, from the perspective of diagnostic application and its biological significance, it is difficult to accept its primacy over cytogenetic value in favorable and unfavorable subgroups and if so, we did not address this issue in our study due to sample size limitation. In intermediate subgroup, particularly in normal karyotype-AML, given the lack of convincing results, it seems unlikely that DNMT3A expression analysis could attract attention in diagnostic workup and risk prediction of AML.
Introduction
As a hematologic malignancy, acute myeloid leukemia (AML) features by an abnormal growth of myeloid white blood cells in bone marrow and consequent perturbation of normal hematopoiesis.Citation1 Rather than this relatively simple definition, AML is notorious for its clinical and molecular heterogeneity.Citation2,Citation3 In the field of molecular heterogeneity, in spite of recurrent alteration in chromosomal and molecular genetics, it has been shown that AML is associated with changes in gene expressions and epigenetic patterns.Citation2–Citation6 It appears that this background molecular heterogeneity brings a distinct leukemic phenotype with it and causes differences in the prognosis and the response to therapy among AML patients.Citation4,Citation7 Despite advances in our understanding of the molecular genetics of AML, most of the patients still fall in the intermediate risk category, without a known cytogenetic or molecular driver. Cytogenetic works well for the patients with an abnormal karyotype and is retaining its place in the clinical settings, but it is difficult to predict the prognosis for the patients with normal karyotype (NK-AML) that comprise approximately 50% of all AML patients.Citation8 In fact, this category of the patients presents variable clinical behaviors and different treatment responses that reflect its heterogeneous molecular background. Despite the advances in molecular genetics of NK-AML and the identification of recurrent genetic abnormalities with functional roles in proliferation and differentiation of myeloid progenitors,Citation3 to date, screening of FLT3, NPM1, and CEBPA mutations have entered in the diagnostic workup of this subgroup with the implication in risk assessment and treatment approach.Citation3 It has been already shown that integrated mutational profiling can improve initial risk stratification of NK-AML patients resulting in better treatment. Therefore, studies to find new marker with prognostic value are still ongoing.Citation3 DNMT3A encodes an enzyme primarily involved in the de novo DNA methylation at CpG sites. For the first time, Yamashita et al.Citation9 have reported DNMT3A mutation in AML. Soon after that, using next-generation sequencing approach, it was shown that mutation in DNMT3A is a recurrent event in AML, with an incidence of approximately 22%.Citation10 In the latter mentioned study, it was also shown that mutations in DNMT3A are more common in NK-AML, with an incidence of approximately 34%, and can be regarded as an independent prognostic factor. It was also noted that DNMT3A mutations, clustered with FLT3, NPM1, and IDH1 mutations, were significantly enriched in patients with M4 and M5 French–American–British (FAB) subtype, and there was a strong selection against core binding factor (CBF) abnormalities.Citation10 Subsequent studies have confirmed these findings and furthermore showed that mutation in DNMT3A is more frequent in elderly patients and associates with higher white blood cell (WBC) and platelet counts and higher MLL5 expression level as compared with patients with wild-type DNMT3A.Citation11–Citation13 In a recently published meta-analysis, these findings has been reaffirmed.Citation14 Despite these congruent opinions on the association of DNMT3A mutation with some distinct biological characteristics and its negative impact on the clinical outcome of AML, there is still a controversy over the incidence of DNMT3A mutation in different population. The frequency of the mutations was found to be 4.1, 9, and 8% in Japanese, Chinese, and Brazilian samples, respectively,Citation9,Citation15,Citation16 and this is different to the frequency of approximately 20%, attributed to European and American populations.Citation10–Citation13 Furthermore, in a recently study, conducted on a sample of Egyptian population, it has been asserted that DNMT3A mutations are highly frequent in AML (28%),Citation17 further highlighted the discrepancy over the occurrence of DNMT3A mutation in different population. Recently it has been shown that hypermethylation at an internal region of the DNMT3A promoter could affect the expression of its variant transcripts. The authors have declared that such silencing event and recurrent mutation of DNMT3A are mutually exclusive.Citation18 To the best of our knowledge, the incidence of DNMT3A mutations in Iranian population has never been studied, and our study could highlight some information as add-on to the results reported on Egyptian patientsCitation17 and together could be considered as a representative of the Middle East population. To further explore, the expression of DNMT3A transcripts of this gene in patients with and without mutations has also been analyzed.
Material and methods
Patients and treatment
Adult patients in the range of 16–60 years, for whom diagnostic peripheral blood (n = 63 with blast ≥30%) or bone marrow (n = 33) samples were available, were included after obtaining Institutional Review Board approval and informing consent in accordance with the Declaration of Helsinki. All patients received the same treatment protocol containing: induction therapy with cytarabine 100–200 mg/m2/d administered as a continuous 7-day infusion plus idarubicin 12 mg/m2/d administered as a bolus intravenous injection on each of the first 3 days of treatment. Patients with complete remission received 3–4 cycles of high-dose cytarabine 3 g/m2 administered in a 3-hour infusion every 12 hour on day 1, 3, and 5 for a total of six doses as consolidation. The patients were classified according to the FABCitation5 and cytogenetic.Citation19 Clinical and biological characteristics of the patients and controls who participated in this study have been given in .
Table 1. Clinical and biological characteristics of the participants
DNA extraction, PCR amplification, and sequencing
Mononuclear cells were separated on Ficoll-Hypaque from samples before treatment. Genomic DNA was extracted using salting-out method. The screening of DNMT3A mutations was performed on six terminal exons 18, 19, 20, 21, 22, and 23 (GenBank reference NM_175629), which in fact included methyltransferase domain as mutational hot spot by PCR. Primer sequences were previously described by Thol et al.Citation12 The total reaction volume of 50 µl contained approximately 100 ng DNA, 0.5 µM of each primer, 0.2 mM of each dNTP, 1 mM MgCl2, and 1 U of Hot Start Taq polymerase and supplied buffers with cycling conditions as follows: 1 cycle for 5 minutes at 94 °C, 35 cycles for 1 minute at 94 °C, 45 seconds at 56 °C, 1 minute at 72 °C, and 1 cycle for 10 minutes at 72 °C.
The quality of the PCR products was verified by electrophoresis on 1.5% agarose gel and all amplicons were sequenced with an ABI 377 automated DNA sequencer (Applied Biosystems, Foster City, California, USA) using the Big Dye terminator V1.1 cycle sequencing kit.
RNA isolation and reverse transcription and qRT-PCR
Total RNA extraction was performed using TRIZOL reagent (Bioneer, Daejeon, Republic of Korea), according to the manufacturer's protocol. After RNA levels, purity, and quality assessment, complementary DNA (cDNA) was synthesized using RevertAid First Strand cDNA Synthesis Kit (Fermentas, Waltham, Massachusetts, USA). In this study, we established qRT-PCR using primers dedicated to the sensitive detection of targeted transcripts. We used ABL1 as reference geneCitation20 (primer pairs are listed in ). Construction of standard curves for quantification of DNMT3A variants and the internal control ABL1, and calculation of DNMT3A variants normalized to ABL1 were performed as previously reported.Citation8 In addition, we determined DNMT3A mRNA levels in leukocytes form peripheral blood samples from 20 healthy age-matched volunteers, although peripheral blood leukocytes are not optimum control for expression analysis in relation to leukemic blasts.
Table 2. Primers for qRT-PCR
Statistical analysis
Kolmogorov–Smirnov test was used to assess the normality of data distribution and Levene's test for equality of variances. For comparing gene expressions between two groups, the independent samples t-test was used for gene expressions with normal distribution, and the non-parametric Mann–Whitney U test for non-normal distributions. Overall survival (OS) was measured from the date of first diagnosis to the date of death from any cause, and relapse-free survival (RFS) was measured from the date of complete remission to the date of relapse or death from any cause, whichever occurred first. Relapse was defined as a reappearance of at least 5% leukemic blasts in a bone marrow (BM) aspirate or new extra-medullary leukemia. OS and RFS curves were calculated using the Kaplan–Meier survival function and comparisons between groups were made using the log-rank test. Multivariate Cox proportional hazard regression analysis was used to investigate independent prognostic factors for OS and RFS. P-value of less than 0.05 was considered statistically significant for all statistical tests. All statistical analysis was performed using SPSS software Version 16.
Results
DNMT3A mutations in patients with de novo AML and its correlation with clinical and biological features
In this study, we performed mutation analysis of last six exons of DNMT3A in 96 patients with newly diagnosed AML. We exclude single-nucleotide polymorphisms and overall, 17 DNMT3A non-synonymous variants were founded in 17 of the 96 patients (17.7%). Of 17 founded mutants, 7 were females (7 out of 40, 17.5%) and 10 were males (10 out of 56, 17.9%) (P = 0.964). The type of mutations was nine missense mutations at position R882 (53%) and eight mutations at other positions (47%), comprising five missense, two nonsense, and a frameshift mutation (). The somatic status of the identified DNMT3A alterations could not be proven owing to the lack of matched DNA from unaffected tissue. Patients with DNMT3A mutations presented with higher age (mean age 48.88 vs. 43.19 years, P = 0.045) and higher WBC counts (mean: 47 200 vs. 27 400, P = 0.01). No differences in platelet count and Hb concentration were observed between patients with and without DNMT3A mutations (P = 0.129 and 0.196, respectively). Regarding cytogenetic, 13 of 17 DNMT3A-mutant AMLs (76.5%) carried an NK-AML, and DNMT3A mutation frequency in NK-AMLs of our study was 36.1% (13 of 36). One mutant belonged to the unfavorable cytogenetic group. We did not find any mutation in patients with favorable cytogenetic group that includes patients with CBF-AML. In accordance with previous reports,Citation10,Citation12,Citation13 DNMT3A mutations were strongly associated with FAB M4/M5 subtypes (13 out of 17, 76.5%, P < 0.001) and 50% of FAB M4/M5 subtypes (13 of 26) carried mutations in DNMT3A (6 out of 14 M4 and 7 out of 12 M5).
Table 3. Characteristics of the mutation pattern of DNMT3A mutants
Relative expression of DNMT3A and its transcript 2
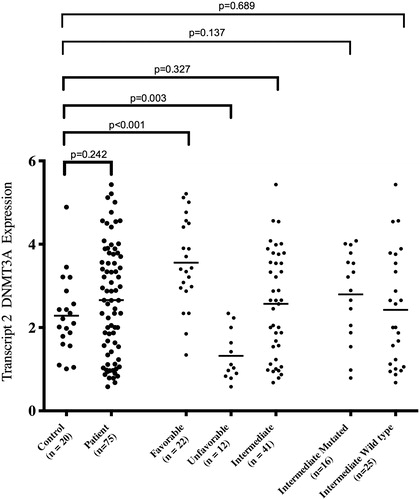
In initial phase, we analyzed DNMT3A expression with a primer pair that matches all available transcripts (named hereafter as total DNMT3A). Our results indicated that total DNMT3A expression did not alter in AML patients compared to normal controls (P = 0.621, ). Then, for more accurate evaluation, we analyzed the expression in three cytogenetic groups. Our results showed that DNMT3A over- and underexpressed in favorable and unfavorable groups, respectively (P = 0.005 and P < 0.001, respectively, ). For intermediate subgroup, we had no different DNMT3A gene expression (P = 0.575, ). In relation to our cytogenetic subgroups and comparing each other, our expression results were in accordance with cytogenetic risk stratification when analyzed in group order (P = 0.001 for favorable vs. intermediate, P < 0.001 for favorable vs. unfavorable, and P < 0.001 for intermediate vs. unfavorable, ). As mentioned above, recently it has been shown that hypermethylation at differentially methylated regions (DMRs) of internal promoter of DNMT3A could reduce expression of its variant transcripts and thereby mimic mutations in DNMT3A. Special emphasis has been on transcript 2 of DNMT3A, in such a way that hypermethylation at DMR2 and DMR3, which seem to be related to the internal promoter region of transcript 2, causes down-regulation of DNMT3A transcript 2.Citation18 To further analysis, we determined transcript 2 expressions in our total patients and three cytogenetic risk groups. We had no different transcript 2 expression in our total cohort as compared with controls (P = 0.242, ). Further focusing, our favorable group showed overexpression (P < 0.001, ) and our unfavorable group showed an underexpression (P = 0.003, ) in terms of the expression of DNMT3A transcript 2. Interestingly, our intermediate group did not show altered transcript 2 expression compared to controls (P = 0.327, ). In addition in our divided intermediate subgroups, to mutated and wild type, the results of the study noted no change in gene expression, compared to normal controls (P = 0.137 and P = 0.689 for mutated and wild type, respectively, ). At this stage and with information we had on hand and according to similar data for the total DNMT3A expression and its transcript 2, we were curious as to the expression status of the other transcripts. We analyzed expression of DNMT3A transcript 1/3 (which cannot be distinguished by distinctive primers; ) and transcript 4 in cytogenetic subgroups of 10 patients each comparing to 10 normal controls using transcript specific primers and interestingly our finding showed no change in expression of these transcripts, although the number of cases studied was rather small (data not shown).
Figure 1. Comparing total DNMT3A gene expression level between patients, cytogenetic risk subgroups, and normal control. It is worth mentioning that normal control RNA was extracted from peripheral blood.
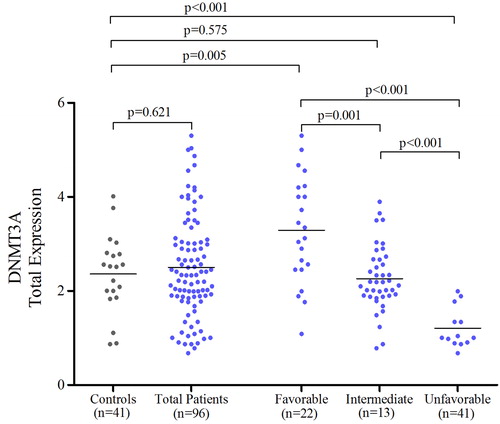
Figure 2. Comparing DNMT3A transcript 2 expression level between total patients, three cytogenetic risk subgroups, intermediate-mutant, intermediate-wild type, and normal control, from left to the right, respectively. It is worth mentioning that normal control RNA was extracted from peripheral blood.
Impact of DNMT3A mutation on clinical outcome
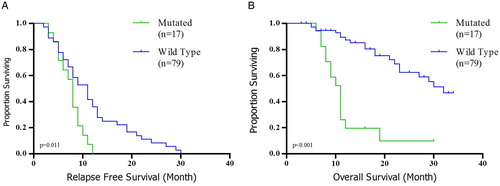
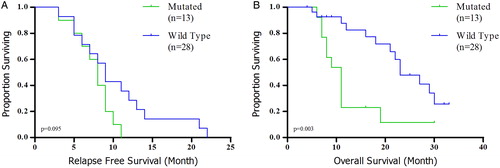
All patients received conventional intensive induction chemotherapy with 33-month follow-up. Patients with the DNMT3A mutation had significantly shorter OS (median: 11 months vs. 29 months, HR: 0.128, 95% CI: 0.055–0.299, P < 0.001) and RFS (median: 8 months vs. 13 months, HR: 0.305, 95% CI: 0.159–0.587, P = 0.011) than those without (A and B). The same was true for OS among intermediate cytogenetic risk group (median: 10 months vs. 19 months, HR: 0.144, 95% CI: 0.046–0.448, P = 0.003, B). We did not have significant relationship for RFS regarding intermediate subgroup (median: 8 months vs. 11 months, HR: 0.363, 95% CI: 0.153–0.864, P = 0.095, A). Multivariate analysis showed that DNMT3A mutant is an independent prognostic indicator for unfavorable OS (HR: 0.131; 95% CI: 0.048–0.355; P < 0.001) and RFS (HR: 0.445; 95% CI: 0.220–0.900; P = 0.024) in the entire when we consider age, WBC count, platelet count, and cytogenetic risk.
Discussion
Mutations in DNMT3A have been reported in patients with de novo AML and with lower frequencies, in other hematological malignancies.Citation21,Citation22 However, the incidence is variable, mainly because of selected cohorts, different ethnical backgrounds or partial/total area of the gene selected for mutation screening. In this study, we have analyzed the mutational frequency of DNMT3A in 96 Iranian de novo AML patients. We sequenced last six exons, which comprise the entire coding region of the methyltransferase domain as mutational hot spot. Our results indicate that DNMT3A mutations have occurred with the frequency of 17.2% in our total cohort and 36.1% in our NK-AMLs. Patients with mutation are associated with distinct clinical and biological features like higher age, WBC count, and M4/M5 FAB type. Besides determining the mutation frequency in this study, we have also shown that DNMT3A mutations have a negative impact on clinical outcome. In our entire cohort, we have had shorter OS and RFS in our DNMT3A mutants as compared with those without such changes. These results are in accordance with previous studies.Citation10,Citation12,Citation13,Citation17 In intermediate risk karyotype subgroup, although we had negative impact of DNMT3A mutation on OS, in relation to RFS, our results are negative and we do not have any relationships. In this case, our finding is in agreement with Thol et al.Citation12 who saw no significant relationship. In this study, for the first time, we have also analyzed the expression of total DNMT3A in conjunction with its mutational screening in our cohort, and our results have shown that the expression did not alter in our total leukemic samples compared to the controls that were different from the results of previous studies emphasizing on DNMT3A overexpression in AML.Citation23 These variable results have been obtained in our study and the mentioned study could be rooted in: (1) features of analyzed patients, such as age and race; (2) different controls used in these two studies. It is important to note that compared to bone marrow samples, used in the mentioned study, we used peripheral blood as normal control that may not be optimized in preference to bone marrow; and (3) most importantly, because of different reference genes, ABL1 in our study and GAPDH in former study. Recall that all housekeeping genes cannot be regarded as a reference gene in AML.Citation20 In addition, for the first time, we analyzed total DNMT3A expression regarding three cytogenetic subgroups. In our favorable subgroup, we have had total DNMT3A overexpression, and for unfavorable subgroup, our results are in opposite direction and we had DNMT3A underexpression, but in intermediate subgroup, total DNMT3A expression did not alter. In relation to total DNMT3A expression and cytogenetic subgroups, although our results are in line with cytogenetic risk prediction, and there is a direct relationship between the expression level of DNMT3A and karyotype-based risk prediction, it should be considered that we have compared the samples with the mature peripheral leukocytes. Nevertheless, these findings highlight the potential prognostic value of DNMT3A expression in risk stratification of leukemic patients and perhaps could be regarded as add-on information to the better risk stratification of favorable and unfavorable subgroups, like what KIT mutation did in favorable risk group.Citation24 We could not take this survey and this is another shortcoming of our study that rooted in small sample size and relativeness of our qRT-PCR results in preference to absolute quantification for each patient and concerning clinical outcomes that could be a proposal to confirm in future studies. In order to narrow down our study to determine the expression status of variant transcripts of DNMT3A, interestingly, we had expression results for DNMT3A transcript 2 similar to that of total DNMT3A. Considering these results and the results of the expression of the other transcript, 1/3 and 4, in general, it can be deduced that although all the transcripts of DNMT3A are expressed in hematopoietic tissues, its transcript 2 expression could alter and this results in total DNMT3A expression changes, as we have seen. Of course, since the gene expression analysis of the other transcript (1/3 and 4) was performed on a small sample size, this statement should be treated with caution. In addition, in a recently published paper, it has been asserted that aberrant hypermethylation of an internal promoter region of DNMT3A occurs in many AML patients, and it is higher in samples without DNMT3A genomic mutation. Special emphasis is on hypermethylation at DMR2 that results in its transcript 2 down-regulation in patients with epimutation in DNMT3A promoter region.Citation18 It has also been mentioned that an even more pronounced down-regulation of transcript 2 was observed in patients with mutations in DNMT3A. To clarify the possible impact of DNMT3A mutation on its expression, we focused on a subset of our cohort. Our study showed different results and as shown in , in intermediate subgroup and comparing DNMT3A mutant and wild type to the controls, we did not have different expression results. This could be due to the fact that in our study, we used a housekeeping gene, ABL1, but in the mentioned study, expression was normalized to DNMT3A own transcript 1/3. Given that regional hypermethylation usually targets the whole CpG islands (CGIs) and shore regions and it does not seem to be restricted to a particular region of promoters.Citation25 This scattered methylation results in different inter-individual expression. It does not seem logical that DNMT3A own other transcript could serve as normalizer, because the expression of these transcripts could also be affected, resulting in high inter-individual variation of expression and a false error to interpret the data, ultimately. However, it should be noted that our overexpression of total DNMT3A and its transcript 2 in favorable subgroup are correlative to founded DNMT3A hypomethylation.Citation18
Conclusion
Till now, several studies with large sample size have stressed on the independent prognostic value of DNMT3A genomic mutations.Citation10,Citation11,Citation13,Citation14 In current studies, this prognostic value has been reaffirmed again in our population. Regarding this general consensus that has been achieved, it seems that the time has come to integrate mutational screening of DNMT3A in risk stratification of AML patients in the settings of integrated mutational profiling. In relation to DNMT3A expression, from the perspective of diagnostic application and its biological significance, it is difficult to accept its primacy over cytogenetic value in favorable and unfavorable subgroups and if so, we did not address this issue in our study due to sample size limitation. In intermediate subgroup, particularly in NK-AML, given the lack of convincing results, it seems unlikely that DNMT3A expression analysis could attract attention in diagnostic workup and risk prediction of AML. By the way, this is an initial study that takes into account DNMT3A expression in AML cytogenetic subgroups, and its results should be confirmed in future large studies.
Acknowledgments
We would like to thank the patients who participated in this study.
Disclaimer statements
Contributors No other contributors.
Funding None.
Conflicts of interest None.
Ethics approval None.
References
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–62.
- Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet 2013;381:484–95.
- Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. (official journal of the American Society of Clinical Oncology) 2011;29:475–86.
- Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood 2013;121:3563–72.
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French–American–British Cooperative Group. Ann Intern Med. 1985;103:620–5.
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010;116:354–65.
- Hajkova H, Markova J, Haskovec C, Sarova I, Fuchs O, Kostecka A, et al. Decreased DNA methylation in acute myeloid leukemia patients with DNMT3A mutations and prognostic implications of DNA methylation. Leuk Res. 2012;36:1128–33.
- Zare-Abdollahi D, Safari S, Movafagh A, Ghadiani M, Riazi-Isfahani S, Omrani MD. Intact expression status of RASSF1A in acute myeloid leukemia. Med Oncol. (Northwood, London, England) 2014;31:770.
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, et al. Array-based genomic resequencing of human leukemia. Oncogene 2010;29:3723–3731.
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–33.
- Ostronoff F, Othus M, Ho PA, Kutny M, Geraghty DE, Petersdorf SH, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: a SWOG report. Leukemia 2013;27:238–41.
- Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. (official journal of the American Society of Clinical Oncology) 2011;29:2889–96.
- Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood 2012;119:5824–31.
- Shivarov V, Gueorguieva R, Stoimenov A, Tiu R. DNMT3A mutation is a poor prognosis biomarker in AML: results of a meta-analysis of 4500 AML patients. Leuk Res. 2013;37:1445–50.
- Pezzi A, Moraes L, Valim V, Amorin B, Melchiades G, Oliveira F, et al. DNMT3A mutations in patients with acute myeloid leukemia in South Brazil. Adv Hematol. 2012;2012:697691.
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–15.
- Ibrahem L, Mahfouz R, Elhelw L, Abdsalam EM, Soliman R. Prognostic significance of DNMT3A mutations in patients with acute myeloid leukemia. Blood Cells Mol Dis. 2015;54:84–9.
- Jost E, Lin Q, Weidner CI, Wilop S, Hoffmann M, Walenda T, et al. Epimutations mimic genomic mutations of DNMT3A in acute myeloid leukemia. Leukemia 2014;28:1227–34.
- Dastugue N, Payen C, Lafage-Pochitaloff M, Bernard P, Leroux D, Huguet-Rigal F, et al. Prognostic significance of karyotype in de novo adult acute myeloid leukemia. The BGMT group. Leukemia 1995;9:1491–8.
- Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using 'real-time' quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) – a Europe against cancer program. Leukemia 2003;17:2474–86.
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012;12:599–612.
- Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 2011;25:1153–8.
- Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 2001;97:1172–9.
- Allen C, Hills RK, Lamb K, Evans C, Tinsley S, Sellar R, et al. The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia 2013;27:1891–901.
- Ushijima T, Okochi-Takada E. Aberrant methylations in cancer cells: where do they come from? Cancer Sci. 2005;96:206–11.