Abstract
Irritable bowel syndrome (IBS) is a chronic disease characterized by complex interactions between genetic predisposition and the environment. Current treatments for IBS are characterized by a highly variable response. Gene variations may result from insertions or deletions, gene rearrangements, splice variants or copy number variants, or, more commonly, from substitutions in the DNA of one (single nucleotide polymorphism [SNPs]) or more than one nucleotide. The objective of this editorial is to review the potential importance of pharmacogenetics in the treatment of IBS based on current evidence.
1. Precision medicine
Precision medicine or individualized medicine is focused on the prevention and treatment strategies that take individual variability into account Citation[1]. Large efforts in precision medicine have impacted treatment of rare disorders, typically monogenic diseases and treatment of cancer. However, there is increased interest to “enable better assessment of disease risk, understanding of disease mechanisms, and prediction of optimal therapy for many more diseases, with the goal of expanding the benefits of precision medicine into myriad aspects of health and health care” Citation[1]. This expanded field includes complex and polygenic conditions taking into consideration the molecular, genomic, cellular, clinical behavioral and environmental parameters with the objective of providing the right drug at the right dose to the right patient. We strongly believe that genotyping and sub-phenotyping IBS will enhance the treatment response and minimize drug-related adverse events.
To identify candidate genes with potential to impact IBS pharmacogenetics, it is necessary to summarize the literature on the association of genetic variations with IBS or with intermediate clinical phenotypes of IBS, such as colonic transit.
2. Genetic variation and IBS phenotype and intermediate phenotype
A genetic predisposition to IBS is supported by twin studies and by the findings from the candidate gene (summarized in Camilleri and Katzka 2012 Citation[2]) that identified associations between SNPs in various genes and IBS symptom phenotypes or quantitative traits (or intermediate phenotypes). The quantitative traits include colonic transit or sensation, intestinal permeability, and mucosal immune activation. The univariately associated SNPs are in genes related to serotoninergic functions (SLC6A4, HT3RA, HT3RE), cannabinoid functions (fatty acid amide hydrolase [FAAH], cannabinoid receptor 1 [CNR1]), bile acid pathways (klotho beta (KLB), G protein-coupled bile acid receptor 1 [GPBAR1]), immune activation (TNFSF15 and IL6 genes), ion channels (SCN9A), barrier or epithelial proteins (NPSR1, TLR9, PRDM1, C11orf30, ORMDL3, CDH1), and HLA2/8 genotype. To date, the TNFSF15 gene has been associated with IBS in three separate cohorts Citation[3-5] even with rigorous corrections for multiple statistical tests.
Recent association studies between large numbers of gene SNPs and IBS symptom phenotype have involved several thousands of patients and have identified other potential genetic associations:
A study of 384 SNPs covering 270 genes in ∼ 1600 people Citation[6] identified association of rs2349775 (NXPH1, associated with neuroticism) in IBS-D and rs17837965 (CDC42, involved in brain development and intestinal stem cell differentiation and proliferation) in IBS-C.
A multi-ethnic, multicenter study of ∼ 6000 IBS patients and including meta-analysis of published data on 16 SNPs in immune candidate genes Citation[7] confirmed only TNFSF15 in IBS, as previously reported Citation[3-5].
In the first GWAS multicenter study of ∼ 8,000 individuals, two genes were identified in association with “risk of IBS”: KDELR2 (KDEL endoplasmic reticulum protein retention receptor 2) and GRID2IP (glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein) Citation[8]. The biological relevance of these two genes in mechanisms of IBS is unknown.
The genetic variations that have been applied in IBS pharmacogenetics are related to serotonin transporter, bile acid pathway and cannabinoid mechanisms. The associations of genetic variations in these three mechanisms and IBS phenotype are introduced here; more exhaustive reviews of genetic associations with IBS have been published elsewhere Citation[2,9].
2.1 Serotonin transporter
A single short allele (44 base-pair deletion in the 5-HT transporter long polymorphic repeat [5-HTTLPR] of the SLC6A4 gene is associated with reduced synthesis of the serotonin (5-HT) transporter protein. Thus, the LS and SS genotypes are associated with less reuptake of 5-HT leaving more of the transmitter in the synapse; conversely, the homozygous L genotype is associated with less synaptic 5-HT. In a meta-analysis of 25 studies involving 3443 patients with IBS Citation[10], the 5-HTTLPR LL genotype of the SLC6A4 gene is associated with IBS-C; this association was only observed in the East Asian cohorts.
2.2 Bile acids
The synthesis, enterohepatic circulation and excretion of bile acids play a major role in the pathophysiology of IBS, particularly in the bowel function. Thus, variants in the genes for proteins involved in the enterohepatic bile acid circulation may modulate colonic transit and are associated with diarrhea. Genetic variation in KLB (Arg728Gln) is associated with diarrhea-predominant IBS (IBS-D) and accelerated colonic transit; the variant KLB gene results in instability of the KLB protein synthesized, resulting in interference with negative feedback by fibroblast growth factor-19 (FGF-19) and increased hepatocyte BA synthesis Citation[11] and diarrhea. Another protein critical for feedback regulation of hepatocyte bile acid synthesis by FGF-19 is the receptor fibroblast growth factor-receptor 4 (FGFR4). FGFR4 genetic variations modulate the association of KLB variation with accelerated colonic transit Citation[11].
2.3 Cannabinoid receptors
Cannabinoid receptors modulate a variety of GI functions, including pain modulation, inflammation, and colonic motility. CNR1 is the gene encoding for cannabinoid-1 receptor (CB1). There was significant association of CNR1 rs806378 genotype with symptom phenotype, with colonic transit in IBS-D (TT having fastest colonic transit) and with sensation rating of gas but not pain during colonic distension Citation[12]. FAAH is the gene encoding fatty acid amide hydrolase, the rate-limiting enzyme for endogenous cannabinoid metabolism. An SNP in the human FAAH gene (C385A) reduces FAAH expression. FAAH CA/AA increases the odds for D-IBS-D, IBS-mixed, and, possibly, chronic abdominal pain relative to healthy volunteers. There was also a significant association of FAAH CA/AA genotype with accelerated colonic transit in D-IBS Citation[13].
3. Pharmacogenetics in IBS
Variations in genes (and their encoded proteins) can alter the outcomes of drug-based therapy through two broad mechanisms: first, variations in inherited genes may affect drug processing as these genes encode transporters, receptors or enzymes involved in metabolic pathways of a drug. Second, variations in the germline or somatic genes that regulate mechanisms involved in the pathophysiology of the gastrointestinal disease also affect drug efficacy.
3.1 Genetic variations in drug metabolism
Cytochrome P450 (CYP) enzymes metabolize endogenous and exogenous compounds. Molecular variations in the genes encoding CYP2D6 have the greatest potential clinical impact in IBS as CYP2D6 metabolism affects tricyclic antidepressants and selective serotonin-reuptake inhibitors. These drugs are used to treat visceral hypersensitivity and pain in IBS. CYP2D6 metabolism can be classified as ultra-rapid, extensive, intermediate or poor, according to the number of functional alleles. Poor metabolizers constitute 5 – 10% among white populations and ∼ 1% among Asian populations. Conversely, gene expression might result in three or more functional CYP2D6 alleles, which accelerate drug metabolism. Although multiple copies of the CYP2D6 gene are relatively infrequent among Northern Europeans, some ethnic groups (e.g., East Africans) have > 10 functional alleles Citation[14].
3.2 Genetic variations in drug targets
Thus far, the variability in the IBS phenotype and treatment response has been applied mostly using pharmacogenetic–pharmacodynamic approach, particularly colonic transit and stool consistency and frequency. Three relevant examples are pharmacogenetics trials that targeted serotonin, bile acids, and cannabinoid pathways. These are summarized briefly.
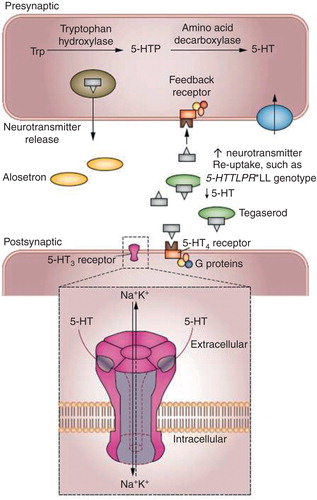
In the presence of the S allele of the 5-HTTLPR polymorphism in the SLC6A4 gene, there is more 5-HT to be competed by a 5-HT3 antagonist used in the treatment of IBS-D, and more 5-HT available to stimulate contractile activity when treatment with a 5-HT4 agonist is indicated clinically, as in the treatment of constipation (). Thus, in individuals carrying 5-HTTLPR LL polymorphism in the SLC6A4 gene, alosetron is more effective in slowing colonic transit in patients with IBS-D Citation[15]. Among patients with IBS-C, carriers of 5-HTTLPR LL are also less responsive to the 5-HT4 agonist, tegaserod Citation[16]. This example supports the benefit of applying a pharmacogenetics approach in a complex disease such as IBS.
Figure 1. Hypothetical explanations for the association of alosetron treatment with slowing of colonic transit in 5-HTTLPR*LL carriers in the SLC6A4 gene with IBS-D, and the worse clinical response to tegaserod in 5-HTTLPR*LL carriers with IBS-C. The 5-HTTLPR*LL carrier status is associated with optimal synthesis of the reuptake serotonin transporter protein, SERT. Therefore, in 5-HTTLPR*LL carriers, less 5-HT remains in the synapse to be competed by alosetron at 5-HT3 receptors, and therefore the same dose of alosetron will inhibit the accelerated colonic transit in IBS-D compared to carriers with an S allele. Conversely, with less 5-HT at the synapse, the same dose of tegaserod will be less effective at stimulating 5-HT4 receptors in 5-HTTLPR*LL carriers, resulting in lower impact of tegaserod on symptoms in constipation-predominant IBS.
Another target with a pharmacogenetics potential is the bile acid pathway. FGFR4 genetic variations are associated with alterations in the colonic transit in response to chenodeoxycholic acid in patients with IBS-C Citation[17] and to colesevelam in IBS-D Citation[18].
The CB1 receptor agonist dronabinol (a synthetic tetrahydrocannabinol derivative) decreased fasting colonic phasic motility and increased colonic compliance in patients with IBS-D. In patients with IBS-D, the CNR1 rs806378 CT/TT genotype was associated with delayed colonic transit in response to dronabinol treatment compared to the CC genotype Citation[19]. Conversely, in patients with IBS-C, dronabinol enhances the proximal colonic motility index in CNR1 rs806378 CC genotype when compared to CT/TT genotype.
4. Conclusion
Although not yet established in the field of IBS, the era of “precision medicine” provides opportunity to conduct pharmacogenetics trials to explore the option of individualizing therapy for IBS. This field will be enhanced with validation of biomarkers or mechanisms relevant to the pathophysiology, in at least a subgroup, of IBS patients.
5. Expert opinion
For the foreseeable future, the impact of pharmacogenetics in IBS will be greatest in modification of dose or choice of therapy based on genetic variations in drug metabolism. For this to occur, it is necessary to include in the electronic health (medical) record of each patient the metabolized status for the most relevant cytochrome P450 enzymes (CYP2D6, 2C19, 3A4). This requires funding or reimbursement of the testing, education of prescribers and dispensing pharmacists, electronic prescription to enhance coordinated care, and an environment where drug interactions or anticipated drug metabolizing problems can be rectified with an alternative therapeutic approach. In the future, Phase III clinical trials should include pivotal genetic variations in drug targets (preferably with minor allele frequencies > 10%) in order to ascertain whether modifiers of response may be related to such genetic variations.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Notes
Bibliography
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793–5
- Camilleri M, Katzka DA. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. Genetic epidemiology and pharmacogenetics in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 2012;302:G1075–84
- Zucchelli M, Camilleri M, Andreasson AN, et al. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut 2011;60:1671–7
- Camilleri M, Carlson P, McKinzie S, et al. Genetic susceptibility to inflammation and colonic transit in lower functional gastrointestinal disorders: preliminary analysis. Neurogastroenterol Motil 2011;23:935–e398
- Swan C, Duroudier NP, Campbell E, et al. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFalpha. Gut 2013;62:985–94
- Wouters MM, Lambrechts D, Knapp M, et al. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut 2014;63:1103–11
- Czogalla B, Schmitteckert S, Houghton LA, et al. A meta-analysis of immunogenetic association studies in irritable bowel syndrome. Neurogastroenterol Motil 2015;27(5):717–27
- Ek WE, Reznichenko A, Ripke S, et al. Exploring the genetics of irritable bowel syndrome: a GWA study in the general population and replication in multinational case-control cohorts. Gut 2014. [Epub ahead of print]
- Camilleri M. Genetics of human gastrointestinal sensation. Neurogastroenterol Motil 2013;25:458–66
- Zhang ZF, Duan ZJ, Wang LX, et al. The serotonin transporter gene polymorphism (5-HTTLPR) and irritable bowel syndrome: a meta-analysis of 25 studies. BMC Gastroenterol 2014;14:23
- Wong B, Camilleri M, Carlson P, et al. A Klothoβ variant mediates protein stability and associates with colon transit in irritable bowel syndrome with diarrhea. Gastroenterology 2011;140:1934–42
- Camilleri M, Kolar GJ, Vazquez-Roque MI, et al. Cannabinoid receptor 1 gene and irritable bowel syndrome: phenotype and quantitative traits. Am J Physiol Gastrointest Liver Physiol 2013;304:G553–60
- Camilleri M, Carlson P, McKinzie S, et al. Genetic variation in endocannabinoid metabolism, gastrointestinal motility, and sensation. Am J Physiol Gastrointest Liver Physiol 2008;294:G13–19
- Camilleri M. The role of pharmacogenetics in nonmalignant gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 2012;9:173–84
- Camilleri M, Atanasova E, Carlson PJ, et al. Serotonin-transporter polymorphism pharmacogenetics in diarrhea-predominant irritable bowel syndrome. Gastroenterology 2002;123:425–32
- Li Y, Nie Y, Xie J, et al. The association of serotonin transporter genetic polymorphisms and irritable bowel syndrome and its influence on tegaserod treatment in Chinese patients. Dig Dis Sci 2007;52:2942–9
- Rao A, Wong B, Camilleri M, et al. Chenodeoxycholate in females with irritable bowel syndrome-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology 2010;139:1549
- Wong BS, Camilleri M, Carlson PJ, et al. Pharmacogenetics of the effects of colesevelam on colonic transit in irritable bowel syndrome with diarrhea. Dig Dis Sci 2012;57:1222–6
- Wong BS, Camilleri M, Eckert D, et al. Randomized pharmacodynamic and pharmacogenetic trial of dronabinol effects on colon transit in irritable bowel syndrome-diarrhea. Neurogastroenterol Motil 2012;24:358–e169
