Abstract
Biomarker research of psychiatric disorders is delayed by symptom pattern-related diagnostic categories that are only distantly associated with biological mechanisms. In neuropsychiatric disorders that have high heritability (schizophrenia, autism, Alzheimer's disease), genomic research led to significant genome-wide association study (GWAS) results by increasing the number of subjects in case–control studies, and thus provided new hypotheses regarding the aetiology of these disorders and possible targets for research of new treatment approaches. In contrast, in moderately heritable psychiatric disorders (anxiety disorders, unipolar major depression), the development of symptoms, in addition to risk genes, is more dependent on the presence of specific environmental risk factors. Thus, controlling for heterogeneity, and not simply increasing the number of subjects, is crucial for further significant psychiatric GWAS findings that warrant the collection of more detailed individual phenotypic data and information about relevant previous environmental exposures. Gene–gene interactions (epistasis) and intermediate phenotypes or psychiatric and somatic co-morbidities, by identifying similar cases within a diagnostic category, could further increase the generally weak effects of individual genes that limit their usefulness as biomarkers. In conclusion, we argue that methods that are suitable to identify biologically more homogeneous subgroups within a given psychiatric disorder are necessary to advance biomarker research.
1. Introduction
Psychiatric disorders impose enormous medical and economic burdens on patients, their families and healthcare providers worldwide. Therefore, there is an unmet need to improve early diagnosis and thus treatment strategies including prevention. However, currently used diagnosis and disease entities in psychiatry are largely based on clinical phenomenology and lack biological validity, which is a major limitation in identifying biomarkers for psychiatric disorders.
In the past decade, genetic research was expected to aid in the definition of psychiatric disease entities themselves by identifying new biological pathways. In some neuropsychiatric disorders that have high heritability, such as schizophrenia, autism and Alzheimer's disease (), increasing the number of persons in case–control studies led to significant genome-wide association study (GWAS) results, and thus provided new hypothesis regarding the aetiology of these disorders and directed research to new treatment approaches Citation[1]. However, we are still missing considerable amount of heritability even in these disorders that might be partially explained by gene–environment interactions or complex interplay between genes Citation[2]. In addition, in psychiatric disorders with moderate heritability, such as most anxiety disorders and unipolar major depressive disorder (MDD; ), environmental factors have an even more important role to determine or moderate genetic effects in the development of symptoms that serve as a basis for the diagnosis. Gene–environment interactions may have different effects: i) in case of biological interaction, the genetic effect depends on the presence or absence of an environmental factor; ii) quantitative interaction results in the same direction of effect in both genotype groups but the effect sizes differ according to the environmental exposure; and iii) qualitative interaction may produce opposite direction of effects in different genotype carriers Citation[2]. The case–control studies that do not contain detailed individual data about relevant previous environmental factors and exposures could have not led to significant GWAS results so far when simply increasing the numbers in these moderately heritable disorders Citation[1]. In a very recent study, four risk loci with shared effects on five major psychiatric disorders (autism spectrum disorder, attention deficit-hyperactive disorder, bipolar disorder, MDD and schizophrenia) were identified after including about 33,000 cases and 28,000 controls in the analysis Citation[3]. Among those, rs2535629 on chromosome 3 showed the smallest p value and thus the strongest association signal, but even with this polymorphism, no association was noted in a large replication dataset of MDD, suggesting that the potential function of this genetic region in MDD is ambiguous Citation[3]. In a study about the possible role of serotonin transporter gene (SLC6A4) promoter polymorphisms (5-HTTLPR) in the development of MDD and low mood, 5-HTTLPR had no significant effect in the whole population. In contrast, it doubled the significant effects of threatening life events (TLEs) when both parameters (5-HTTLPR and TLE) were included in the model Citation[4]. A recent large meta-analysis, including > 40,000 subjects, provided robust evidence that 5-HTTLPR moderates the relationship between life stresses and depression Citation[5]. Thus, despite the methodological and statistical difficulties, in the future the inclusion of environmental factors in datasets and analysis is crucial for psychiatric disorders with moderate heritability (e.g., MDD, anxiety disorders, drug/alcohol/tobacco dependence), and even in those with high heritability, the inclusion of environmental factors will shed light on further genetic components not discovered yet.
Table 1. Heritability of psychiatric disorders and summary of relevant genetic studies.
However, there is a general limitation of GWAS studies, namely the small effect size of genes on phenotypic variance. Even in the few variants that markedly change protein structure or function, the association with complex psychiatric phenomena, such as diagnostic categories, is weak, making it difficult to use them in personalised diagnosis. One possible approach to increase our understanding of genetic effects is to investigate gene–gene interactions or epistasis, where the combination of two or more polymorphisms results in marked biological differences among the groups. Double knock-out animal studies demonstrated the existence of additive genetic effects, where the measured phenotype became more severe with increasing number of risk alleles, but also supported the importance of non-additive genetic interactions in which case new phenotypes occurred Citation[6]. Similar evidence has been described in humans between genes of brain-derived neurotrophic factor (BDNF) and 5-HTTLPR on a key brain circuitry relevant in depression Citation[7] and between promoter polymorphisms of cannabinoid receptor type 1 (CB1) gene (CNR1) and SLC6A4 on anxiety trait and temperament Citation[8]. These types of interactions may serve to identify groups of patients with highly increased or reduced risk for certain drug effects or environmental factors Citation[9]. In addition, gene–gene interaction studies pointed out that these effects are apparent when continuous traits are investigated and rarely, if ever, apparent at a diagnosis level Citation[6,9].
Biomarkers are by definition measurable indicators of normal or pathological biological processes and may be influenced by environmental factors, such as drug treatment Citation[10]. By measuring specific biomarkers (), we can gather information about the so-called intermediate phenotypes that in neuropsychiatry represents neurobiological processes with a causal role in the disease pathway Citation[11]. Although intermediate phenotypes are interpreted in several ways, we adopted the definition of Meyer–Lindenberg and Weinberger Citation[12] and presumed that intermediate phenotypes are genetically determined traits (). Thus, different intermediate phenotypes, and those specific biomarkers that characterise them, can be crucial to identify otherwise weak or unobservable genetic effects on complex phenotypes of interest (e.g., diagnosis or therapeutic outcome) by reducing the heterogeneity. It is important to note that the terms intermediate phenotype and endophenotype are not interchangeable because endophenotypes do not necessarily mediate genetic effects to disorders Citation[13]. As an example of the usefulness of different intermediate phenotypes, we investigated the effect of the CREB1–BDNF–NTRK2 pathway on depression. Using rumination, a well-known cognitive intermediate phenotype for depression, the major alleles of the CREB1 and BDNF genes promoted the development of depression. However, in case of history of childhood abuse, the minor alleles of the same genes were risk factors for depression most likely by altering the function of the hypothalamic–pituitary–adrenal (HPA) axis, which is an alternative intermediate phenotype in stress-related disorders Citation[14]. Similarly, the 5-HTTLPR short allele increases the risk of depression and anxiety in the presence of negative life events by increasing amygdala activity following stressful events, and predicts poorer antidepressant response (). But 5-HTTLPR short allele is also associated with better performance in several cognitive tasks and increased social conformity (with similar results in non-human primates) and with a more advantageous effect of psychotherapy presumably because it alters the development of limbic–cortical neuronal connections Citation[15].
Figure 1. The complex interaction of genes, environmental factors and intermediate phenotypes in biomarker research to predict different endophenotypes and outcome variables. A given gene set represents a genetic pathway that influences a specific biological process. Arcs between gene sets represent possible interactions (gene × gene, epistasis). Arrows originated from gene sets depict genetic effects on intermediate phenotypes. Intermediate phenotypes, which denote genetically determined neurobiological processes with causal role in the disease pathway, could be identified as biomarkers themselves but usually difficult to measure directly. However, they could be characterised by using different selected more easily measurable biomarkers (boxes in the middle). We suggest that those specific biomarkers are important and useful for personalised treatment in neuropsychiatric diseases that are informative for intermediate phenotypes influenced by environmental factors and could be used in the clinical diagnosis, outcome, therapeutic and/or side effect of drugs. Arcs between biomarkers represent possible combination of different biomarkers that provide relevant information to predict diagnosis, outcome or drug response/side effects. The environmental factors may cause epigenetic changes (e.g., up- or down-regulation in the expression of particular genes) or direct alterations in the measurable biomarker (e.g., exposure to a neurotoxin or brain trauma that disables or disconnects functional brain areas).
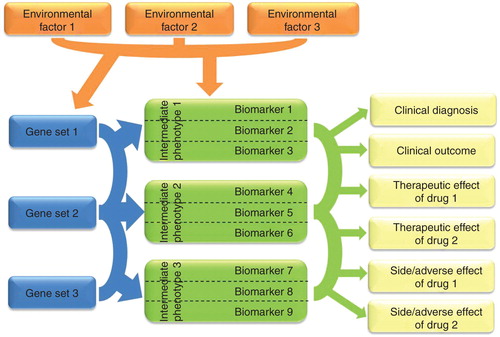
Figure 2. An example of usefulness of brain imaging intermediate phenotypes to identify risk gene and biomarker to treatment response for depression. Life stresses (e.g., watching fearful or sad faces) activate the amygdalae in the brain. The presence of a risk allele within the serotonin transporter gene (SLC6A4, 5-HTTLPR short allele) is associated with increased amygdala activation and with altered connectivity between the amygdalae and prefrontal cortical (PFC) areas compared to non-risk allele carriers during stressful situations. These biomarkers (amygdala activity and PFC–amygdala connectivity), which could be reliably measured by fMRI using selected neuropsychological tasks, provide information how the brain processes negative emotions. Impaired negative emotion processing is an intermediate phenotype that increases the risk of depression and predicts poorer antidepressant response.
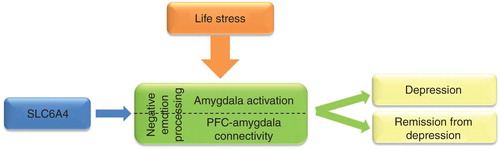
For psychiatric disorders, the aimed intermediate phenotypes are mostly present in the brain, although some exemptions exist (e.g., certain adverse reactions of drugs). Epigenetic changes, which alter gene expressions depending on the environmental exposures, are therefore tissue and cell specific on the periphery; they may be fairly different from those that happen in the central nervous system that is in the focus of interest. Even in the brain, adjacent neurons and nuclei may react differently to a given action or environmental stimulus Citation[16]. Thus, results from the analysis of peripheral biomarkers are difficult and need caution to interpret. Because local invasive sampling of the brain is not possible, the best alternatives are imaging techniques. Functional magnetic resonance brain imaging (fMRI) with carefully selected psychological tasks and positron emission tomography (PET) using specific tracers demonstrated that the functional activity of neural circuits, related to psychiatric phenotypes, are under the influence of genes and environment. For example, one of the most replicated finding is the influence of serotonin transporter gene (and life stresses) on negative emotion processing that can be quantified by measuring amygdala activity and connectivity between amygdala and prefrontal cortex () Citation[12]. Further studies demonstrated that these biomarkers are related to depression and remission from the depressed state Citation[17]. However, it has to be emphasised that these brain mechanisms are not unique for specific psychiatric disorders which is in line with the evidence that current diagnostic boundaries are artificial and high co-morbidities exist between different diagnostic categories.
An emerging new field of biomarker research is to identify not only psychiatric but somatic also co-morbidities that can have shared pathomechanism. The aim of the so-called network medicine is to utilise molecular relationships between disorders that are strikingly different at a phenotypic level Citation[18]. A good example is the high co-morbidity between depression and type 2 diabetes that initiated research into the effects of insulin in neurotransmission and neuroinflammation, and its potential therapeutic use Citation[19].
Finally, to identify biomarkers we have to carefully take into account the phenotype we would like to predict as the complex behavioural phenotypes, such as depression and schizophrenia, or their modification by a drug, which are dependent on several neurobiological processes () Citation[20]. Markedly different or only partially overlapping genes and biological processes are involved in the development of symptoms that lead to the diagnosis; other pathways may play a role in the disease outcome, again others in the therapeutic effects, and further processes in the side and/or adverse effects of a given drug. In the development of tolerance, further mechanisms may be involved. In addition, if we have a drug that works by a different mechanism of action, binds to different receptors and has a different chemical structure, completely new biomarkers may be useful (). For example, biomarkers that could be influenced by both genetic and environmental factors and are substrates of an actual biological process, which is presumably involved in the pathomechanism, may be seemingly perfect for the detection of a drug or other treatment effect. However, in the case of bapineuzumab treatment, a monoclonal antibody against amyloid-β, the PET marker (11C-PiB) of cortical fibrillar amyloid-β load in vivo provided evidence for the disappearance of the protein after 78 weeks. Unfortunately, the mental decline of the patients was not affected, suggesting that there was a dissociation between the effects of the drug observed by the promising marker and the symptoms targeted Citation[21].
2. Expert opinion
Now it is widely accepted that psychiatric diagnoses and disease entities classified by clinical phenomenology do not reflect pathogenic biological mechanisms. In case of highly heritable psychiatric disorders, such as schizophrenia, autism and Alzheimer's disease, GWASs were able to identify common genetic risk variants by increasing the number of subjects in case–control studies, and thus we gained new insight into the possible aetiology of these disorders that triggered the research of new treatment approaches. However, in psychiatric disorders with moderate heritability (most anxiety disorders and unipolar MDD), this method has not provided significant GWAS results most likely because environmental factors interact with genes to produce the symptoms that serve as a basis for diagnosis. Variable environmental exposure could introduce considerable heterogeneity, and thus case–control studies that do not contain detailed individual data about relevant previous environmental factors and exposures did not, and very likely could not, lead to significant GWAS results simply by increasing the numbers. In addition, analysis of gene–environment interaction could have a major role to improve our understanding of highly heritable neuropsychiatric disorders as well. Another possible approach to increase the generally weak effects of individual genes in psychiatry is to investigate gene–gene interactions or epistasis. A different strategy could be the introduction of the so-called intermediate phenotypes that are measurable, genetically determined and environment-dependent neurochemical, anatomical, physiological or psychological phenotypes with a causal role in the disease pathway. Since these are mainly related to brain function, functional brain imaging can provide useful intermediate phenotypes for further genetic and biomarker studies. It is also important to investigate psychiatric and somatic co-morbidities with shared pathomechanisms to identify potential disease pathways. Because disease entities are not based on biological processes in psychiatry, another key point is that symptoms, disease outcomes, therapeutics or side effects of drugs are dependent on several, mostly different or only partially overlapping genes and biological processes.
In conclusion, the major challenge is to develop reliable methods and identify useful biomarkers to minimise heterogeneity of patients within different psychiatric diagnostic categories and get closer to the specific neurobiological processes typical for the given, relatively homogeneous subgroup. Therefore, to extend genetic studies to a more detailed intermediate phenotypes and environmental exposures will be a key strategy for future research to identify new treatment targets and the best biomarkers for the different psychiatric phenotypes.
Declaration of interest
The research of the authors were supported by the Sixth Framework Programme of the European Union (LSHM-CT-2004-503474), the National Institute for Health Research Manchester Biomedical Research Centre (HRF T03298/2000) and the Hungarian Ministry of Health RG (318-041-2009 and TAMOP-4.2.1.B-09/1/KMR-2010-0001).
Bibliography
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012;13(8):537-51
- Thomas D. Gene–environment-wide association studies: emerging approaches. Nat Rev Genet 2010;11(4):259-72
- Smoller JW, Craddock N, Kendler K, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013;381(9875):1371-9
- Lazary J, Lazary A, Gonda X, et al. New evidence for the association of the serotonin transporter gene (SLC6A4) haplotypes, threatening life events, and depressive phenotype. Biol Psychiatry 2008;64(6):498-504
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry 2011;68(5):444-54
- Murphy DL, Uhl GR, Holmes A, et al. Experimental gene interaction studies with SERT mutant mice as models for human polygenic and epistatic traits and disorders. Genes Brain Behav 2003;2(6):350-64
- Pezawas L, Meyer-Lindenberg A, Goldman AL, et al. Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Mol Psychiatry 2008;13(7):709-16
- Lazary J, Lazary A, Gonda X, et al. Promoter variants of the cannabinoid receptor 1 gene (CNR1) in interaction with 5-HTTLPR affect the anxious phenotype. American journal of medical genetics Part B. Neuropsychiat Genet 2009;150B(8):1118-27
- Lazary J, Juhasz G, Hunyady L, Bagdy G. Personalized medicine can pave the way for the safe use of CB(1) receptor antagonists. Trends Pharmacol Sci 2011;32(5):270-80
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001;69(3):89-95
- Lenzenweger MF. Endophenotype, intermediate phenotype, biomarker: definitions, concept comparisons, clarifications. Depress Anxiety 2013;30(3):185-9
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci 2006;7(10):818-27
- Walters JT, Owen MJ. Endophenotypes in psychiatric genetics. Mol Psychiatry 2007;12(10):886-90
- Juhasz G, Dunham JS, McKie S, et al. The CREB1-BDNF-NTRK2 pathway in depression: multiple gene-cognition-environment interactions. Biol Psychiatry 2011;69(8):762-71
- Bagdy G, Juhasz G, Gonda X. A new clinical evidence-based gene-environment interaction model of depression. Neuropsychopharmacol Hung 2012;14(4):213-20
- Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology 2013;38(1):124-37
- Arnone D, McKie S, Elliott R, et al. Increased amygdala responses to sad but not fearful faces in major depression: relation to mood state and pharmacological treatment. Am J Psychiatry 2012;169(8):841-50
- Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12(1):56-68
- Cline BH, Steinbusch HW, Malin D, et al. The neuronal insulin sensitizer dicholine succinate reduces stress-induced depressive traits and memory deficit: possible role of insulin-like growth factor 2. BMC Neurosci 2012;13:110
- Uher R, Tansey KE, Malki K, Perlis RH. Biomarkers predicting treatment outcome in depression: what is clinically significant? Pharmacogenomics 2012;13(2):233-40
- Rinne JO, Brooks DJ, Rossor MN, et al. 11C-PiB PET assessment of change in fibrillar amyloid-beta load in patients with Alzheimer's disease treated with bapineuzumab: a phase 2, double-blind, placebo-controlled, ascending-dose study. Lancet Neurol 2010;9(4):363-72
- Uher R. The role of genetic variation in the causation of mental illness: an evolution-informed framework. Mol Psychiatry 2009;14(12):1072-82