This collagen-based structure by Julian Voss-Andreae is to be installed in the City of South San Francisco’s Orange Memorial Park Sculpture Garden.

The Virginia Bioinformatics Institutes (VBI) has recently launched the VBI Microbial Database (VMD). The work described in Nucleic Acids Research was completed by Brett Tyler, VBI Research Professor and Professor of plant pathology, physiology and weed science at Virginia Tech. (VA, USA), and VBI researchers Sucheta Tripathy, Varun Pandey, Bing Fang and Fidel Salas. The VMD will contain genome sequence and community annotation data, toolkits (an additional resource for users to perform a variety of sequence analysis jobs), and resources to perform complex queries of biological information for a range of microbial pathogens. The aim of the VMD is to make recently completed genome sequences and powerful analytical tools publicly available in one integrated resource. A browser has been created to enable users to view interconnected genome sequence data and annotation pages for each sequence within the database. The community annotation interface allows registered members to add or edit annotations.
The VMD hosts data from a range of plant pathogenic oomycetes, fungi and bacteria, primarily those under study at the VBI. In collaboration with the US Department of Energy Joint Genome Institute, the database currently contains details for Phytophthora ramorum (also known as sudden oak death, a fungal pathogen that poses a serious threat to oak trees in California and Oregon, USA) and Phytophthora sojae (sister pathogen to P. ramorum, which causes severe damage to soybean crops), but is expected to expand. In 2006, the genome sequences for the fungal pathogen Alternaria brassicicola and the oomycete pathogen Hyaloperonospora parasitica will be added to the VMD. A. brassicicola and H. parasitica can both infect Arabidopsis, a commonly used model plant. Support for proteomic and microarray data will also be added, and these data will be linked to the functional genomic data and genome sequences.
The VMD can be accessed at http://phytophthora.vbi.vt.edu.
Analysis of the molecular machines in Saccharomyces cerevisiae completed
Researchers in Germany have completed the analysis of the molecular machines in Saccharomyces cerevisiae. The study from the biotechnology company Cellzome, in collaboration with the European Molecular Biology Laboratory (EMBL), which has appeared in the online edition of Nature, extracted complete protein complexes from cells using tandem affinity purification. Mass spectrometry and bioinformatics where then used to investigate the entire protein household. A total of 257 previously unobserved machines were observed as well as revealing new components of known complexes.
“To carry out their tasks, most proteins work in dynamic complexes that may contain dozens of molecules,” said Giulio Superti-Furga, who launched the project at Cellzome. “If you think of the cell as a factory floor, up to now, we’ve known some of the components of a fraction of the machines. That has seriously limited what we know about how cells work. This study gives us a nearly complete parts list of all the machines, and it goes beyond that to tell us how they populate the cell and partition tasks among themselves.”
During different stages of the cell cycle, and in response to environmental challenges, molecular machines are continually dismantled and reassembled. However, these machines are not built-up from their basic counterparts each time.
“We’ve discovered that the reality is different,” says Anne-Claude Gavin, former Director of Molecular and Cell Biology at Cellzome, and currently a team leader at EMBL. “Cells use a mixed strategy of prefabricating core elements of machines and then synthesizing additional, snap-on molecules that give each machine a precise function. That provides an economic way to diversify biological processes and also to control them.”
Therefore, the cell can respond to stimuli quickly by either producing a few parts to activate or tune the machine, or block molecule production to halt the machine. To reveal these complex interactions, Patrick Aloy and Rob Russell at EMBL used computer techniques. Researchers at EMBL also developed new computational techniques during the study, which enabled them to reveal the dynamic nature of protein complexes.
“This is the most complete set of protein complexes available and probably the set with the highest quality,” says Aloy. “Most proteomics studies in the past have shown whether molecules interact or not, in a ‘yes/no’ way. The completeness of these data lets us see how likely any particular molecule is to bind to another. By combining such measurements for all the proteins in the cell, we discovered new complexes and revealed their modular nature.”
“Investigating protein complexes has always posed a tricky problem – they’re too small to be studied by microscopes, and generally too large to be studied by techniques like x-ray crystallography,” said Russell. “But they play such a crucial role in the cell that we need to fill in this gap. There’s still a huge amount to be learned from this data and from the methods we are developing to combine computational and biochemical investigations of the cell.”
Peer Bork, Head of the Structural and Computational Biology Unit at EMBL, and one of the authors of the paper, said, “Ultimately, we hope to achieve a ‘molecular anatomy’ that takes us from the level of the entire cell to the much deeper level of all the molecules and atoms that make it up.”
The study of S. cerevisiae (baker’s yeast), which is one of the most important model organisms because it is evolutionary related to the cells of animals and humans, may provide clues to human molecular machinery arrangements, and could help aid drug discovery and development. “The same principles discovered here in yeast apply to human cells,” said Gitte Neubauer, Vice President at Cellzome. “Drug targets and pathologically relevant proteins are parts of machines and pathways.”
First library of publicly available breast cancer proteins launched by Harvard Medical School
Havard Medical School (MA, USA) investigators have announced in the Journal of Proteome Research that they have created the first publicly available library of reliably expressible proteins of a human disease, breast cancer. The library, named Breast Cancer 1000 (BC1000), is a compilation of cDNA associated with breast cancer. cDNAs were selected for inclusion in the library through three routes:
| • | Breast cancer research experts in Boston, USA, suggested the first 200 genes | ||||
| • | 50 genes that are overexpressed in ductal carcinoma (a form of breast cancer) were included | ||||
| • | A search of the MEDLINE database using MedGene (a literature-mining software application that was developed at the Harvard Institute of Proteomics) identified the remaining 1050 genes | ||||
A subset of the 1300 cDNAs in the library were expressed in a model system that mimicked human breast cells. This enabled the study of the roles these proteins may play in the development of breast cancer. This approach also enabled the identification of potentially novel functions of both well-known and less conspicuous breast cancer-associated proteins.
“The process of carcinogenesis is complex and involves the activation of many different cellular programs,” explained Joan Brugge, Chair, Havard Medical School Department of Cell Biology, and co-principal investigator of BC1000. “A significant limitation for breast cancer research has been the inability to distinguish whether certain proteins that are altered in breast tumor cells are the cause or the effect of conversion of normal breast cells to malignancy. The systematic approach that we’ve enabled and demonstrated will allow researchers to track cancer-causing proteins in simulated environments, with the goal of learning how to impede them.”
Joshua LaBaer, Director of the Harvard Institute of Proteomics and co-principal investigator, feels that, “The availability of this collection will enable pilot experimentation and accelerate the development of faster techniques for studying breast cancer in a mammalian setting... To advance breast cancer research quickly, we are making the BC1000 library publicly available.”
“Drug design teams in the pharmaceutical industry traditionally have not used proteomics approaches to screen for potential targets, primarily because systematic proteomic tools are in their infancy,” said Steven Carr, who was not part of this research team, but leads the Proteomics group at the Broad Institute (of Harvard University and Massachusetts Institute of Technology). “While this work is highly in vitro and needs further validation, the tools and approaches demonstrated in this study show a potentially valuable screening tool for drug companies, primarily as a means to triage for novel targets to design drugs around,” said Carr. “This study helps lay the groundwork for new and refined proteomics tools for cancer and other diseases.”
The BC1000 library offers a number of advantages over the previous random, nonspecific pooled cDNA libraries:
| • | Each clone’s identity is known | ||||
| • | The quality of each clone is ensured (i.e., full-length and lack mutations) | ||||
| • | Complex phenotypic assays are feasible because there is not the same redundancy that is found in pooled cDNA libraries | ||||
| • | Rare cDNAs are more likely to be represented | ||||
The importance of a breast cancer protein library is of particular interest due to the relatively high incidence of this cancer; only lung cancer causes more deaths in women. The American Cancer Society approximated that 211,240 new cases of breast cancer would be diagnosed among women in 2005. Furthermore, 58,490 cases of noninvasive breast cancer were also estimated.
“The work to isolate, sequence and validate the BC1000 cDNAs was an immense undertaking, with multiple parties involved,” said LaBaer. “While the library covers a broad spectrum of breast cancer-related genes, it is not all inclusive,” he continued. “The addition of new genes to this collection, including genes more recently linked to breast cancer and genes more difficult to clone, is an ongoing effort.”
The BC1000 library can be viewed from the Harvard Institute of Proteomics website (www.hip.harvard.edu).
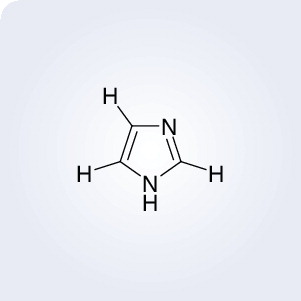
Imidazole ‘on/off’ switch used to study Src’s role in cancer
Johns Hopkins (MD, USA) researchers have found a new way to study the tyrosine kinase Src, a protein with an important, but incompletely understood, role in cancer. The researchers developed a mutated version of the Src protein, which was incorporated into live animal cells. The mutated protein was inactive, but could be activated by imidazole . Imidazole could fit into a pocket in the mutated Src protein structure, which reinstated the Src activity. Removal of imidazole quickly rendered the protein inactive again.
“This strategy provided a controlled environment to study Src,” said Philip A Cole, Director of the Department of Pharmacology and Molecular Sciences at The Johns Hopkins University School of Medicine. “This helped us uncover some new and unexpected insights into how the cancer protein creates its havoc, as well as new treatment leads.” For example, their model demonstrated that Src interacts with CrkL, a signaling protein that had not previously been known to be a target for Src. Furthermore, they found direct evidence that Src activates mitogen-activated protein kinase (MAPK) pathways. The MAPK pathways help transfer signals from growth factors, which are molecules that play a role in the development of cancer cells. The role of Src in the MAPK pathways was previously debated.
“Understanding the functions of different proteins in normal states and disease states is crucial for treatment development because it can help identify new therapeutic targets,” said Cole. “For example, Gleevec® [imatinib], which is used to treat gastrointestinal stromal tumors and chronic myeloid leukemia, is the most successful magic bullet against cancer in many years, and works by blocking tyrosine kinase activity.”
Cole and his coworkers plan to examine the role of Src in cancer in more detail using their new model, and a drug screen could also be developed. Cole thinks that other diseases involving tyrosine kinases, such as the immune system disorder agammaglobulinemia, could also be studied using his technique.
New blood test, the BT Test™, for breast cancer enters clinical trials
Biomarker Technologies, LLC have launched a clinical study for their new blood test, the BT Test™, which detects breast cancer. The BT Test is a blood diagnostic that detects a number of cancer-related biomarkers and can thus detect breast cancer at the molecular level. The study will search for cancer-related biomarkers that have measurably different concentrations in breast cancer patients. A total of 430 women referred for biopsy, 125 women referred for other types of cancer and 300 healthy women will be tested during the study.
Previous studies have demonstrated that the accuracy of the BT Test can significantly exceed that of mammograms. “We expect the results of this clinical study to demonstrate an even higher level of accuracy than either film or digital mammography,” said William Gartner, Chief Executive Officer and President of Biomarker Technologies. “With this greatly improved diagnostic accuracy, the BT Test will ultimately become a critical tool in detecting breast cancer without the discomfort and inconvenience of a mammogram.”
The BT range can be used to study a greater range of patient ages, is more sensitive and is easier to use, compared with the mammogram. The greater sensitivity can also increase the number of early detections, which can lead to more effective therapy.
“This clinical study will move the BT Test forward in its [US] FDA approval as first a supplementary and then a stand-alone screening tool for early breast cancer detection,” said Gartner. “With fewer false-negative and false-positive diagnoses, this cost-effective blood test may not only offer vastly superior early detection capabilities in routine examinations, but may also help patients avoid unnecessary needle biopsies.”
The study will also examine the BT Test in conjunction with the Riboflavin Carrier Protein biomarker from RCP Diagnostics, LLC. This biomarker may further enhance the sensitivity of the BT Test. The study should take 4 months to complete following the collection of blood samples in early March 2006.
Fluorescent Amplification Catalyzed by T7-polymerase Technique could aid early detection of breast cancers
Researchers at the University of Pennsylvania School of Medicine (PA, USA) have developed a new method to detect small amounts of proteins in blood. Fluorescent Amplification Catalyzed by T7-polymerase Technique (FACTT) is 5 orders of magnitude (100,000 times) more sensitive than enzyme-linked immunosorbent assay (ELISA; the current methodology). This sensitive technique will enable the detection of low-abundance molecules associated with cancers (e.g., breast cancer), Alzheimer’s disease, prion diseases and, possibly, psychiatric diseases. The technique will also make these detections more accurate compared with current techniques.
ELISA uses enzymes linked to antibodies or antigens as markers for specific proteins. The FACTT technology uses a different enzyme amplification system, and thus requires fewer sample protein molecules to derive quantitative signals.
“The current ELISA tests can only detect proteins when they are in high abundance,” said Hongtao Zhang, Research Specialist. “But the problem is that many of the functional proteins – those that have a role in determining your health – exist in very low amounts until diseases are apparent and cannot be detected or measured at early stages of medical pathology. It was important to develop a technique that can detect these rare molecules to detect abnormalities at an early stage.”
Mark I Greene, the John Eckman Professor of Medical Science, and coworkers used their new method to test for Her2/neu and compared their results against data obtained through ELISA. Her2/neu is a low-abundance protein that is overexpressed in over 30% of breast, ovarian and pancreatic tumors. Her2/neu overexpression can be determined using blood samples, because part of the Her2/neu molecule is shed by the tumor cells into the bloodstream of breast cancer patients. Higher blood concentrations of Her2/neu correlate with reduced response to chemotherapy and shorter survival times after relapse.
FACTT was found to detect much smaller tumors in animal models, compared with ELISA. FACTT also detected the tumors earlier than ELISA. “The critical issue arises when women are diagnosed with early breast cancer,” said Greene. “They often have a lumpectomy and are sometimes treated with radiation or chemotherapy, but despite this conventional therapy the cancer still can occasionally reoccur.” FACTT may be able to recognize the early phases of tumor emergence, thereby enabling the earlier, and possibly more successful, treatment of these cancers.
“The technology is remarkably adaptable to any protein, and can be performed in an automated format,” said Greene. The use of a robotic system would enable tiny blood samples to be used to screen for many rare disease-causing proteins. “It is even possible that one could screen for multiple diseases at the same time and produce a precise accounting of whether disease-causing molecules are present at an early time when disease can be readily treated,” Greene added.
Greene, Zhang, Xin Cheng (Research Investigator) and Mark Richter (a Research Technician in Greene’s laboratory) reported their findings in Nature Medicine.
Syngene announces the launch of two new automated imagers
Syngene has recently announced the launch of two new automated imagers, Dyversity and G:BOX Chemi XT16 . The first of these, Dyversity, is a fully automated 2D gel image-capture system, which was designed with the aim of saving time when producing 2D protein gel images. Dyversity comprises a light-tight darkroom that contains a high-resolution, 16-bit charge-coupled device camera. This 90-µm resolution camera has the fastest capture times per channel for Cy™ dyes of any system of its type.
There are a variety of precision-made filters, UV and white lighting modules that can be fitted to Dyversity. These enable various protein stains, such as Coomassie Blue, Deep Purple™, Pro-Q® Diamond, silver stain and SYPRO® Ruby, to be imaged. A Cy dye lighting platform is also available. Results produced from Dyversity can be rapidly analyzed using Syngene’s new Dymension software.
Laura Sullivan, Syngene’s Divisional Manager said, “We are very proud of Dyversity because it features unrivalled versatility, which means scientists can choose to use it for a variety of gel imaging applications. The system is so flexible that users can easily upgrade it for different applications, as well as add the filters and lighting they need to image new commercial protein dyes as they become available. This future proofing coupled with the accuracy of its imaging results makes Dyversity the perfect alternative to laser-based scanners.”
Syngene’s second new imager, G:BOX Chemi XT16, is an automated chemiluminescence image analyzer. The camera within this imager is an ultra cooled, high-resolution camera, which can automatically separate close band and spot images. Similar to Dyversity, G:BOX Chemi XT16 contains a 16-bit camera within its own light-tight darkroom. This camera has 4-megapixel resolution in a 2048 × 2048 pixel format. Sullivan explained, “With a high-resolution camera of 4 megapixels, the G:BOX Chemi XT16 is the most sensitive system currently available. Additionally, because the camera is ultra cooled, you can expose blots for as long as you like and still obtain perfect chemiluminescent results with hardly any background every time.”
GeneTools, Syngene’s premier image analysis software, automates tasks, such as molecular weight calculations, band or spot matching and multilayer gel analysis. This automation saves hours of tedious manual analysis. Again, similar to Dyversity, a range of light converters (e.g., a blue light converter, dual UV Epi or blue Epi lighting) can be used to view a wider range of images. Sullivan feels that, “For scientists looking at unrivalled performance with their chemiluminescent imaging, then the G:BOX Chemi XT16 is the best choice.”
Waters Corp. strenghtened by acquisition of VICAM™ technology and collaboration with Thermo Electron Corp.
Waters Corp. have purchased the food safety technology business and associated net assets of VICAM™. VICAM produce single-use food kits that are designed to test for mycotoxin and microbiological organisms in foods. These tests have a high consumer demand due to the requirement for contaminant-free supplies of foods. Not only are mytotoxins a common concern (in 2004, the Annual Report of the European Commissions’ Rapid Alert System for Food and Feed reported that 44% of Information Notifications by identified risk were for mycotoxin contamination in food imported to, or grown within, the European Union), but they are also one of the most carcinogenic compounds currently known.
VICAM immunoaffinity columns isolate contaminants, which are then analyzed in a laboratory using sensitive analysis methods, such as fluorometer measurement, high-performance liquid chromatography, Ultra Performance LC™ (UPLC) and mass spectrometry.
“What initially attracted Waters to VICAM was its leadership position in this important segment of the food testing business, coupled with its strong customer commitment and their focus upon creating relationships with relevant regulatory agencies,” explained Mark Baynham, Director of Chemistry and Consumables Marketing, Waters Corp.
“VICAM is extremely pleased to be aligned with a company that will help it extend its technology platform across many more market opportunities,” said Marjorie Radlo, Executive Vice President, VICAM. “With our already strong market presence in the food safety arena, VICAM’s proprietary products linked to Waters leading separations and detection capability will enable food technologists to further meet the demands of new regulations leading to a safer food supply.”
Waters Corp. is further strengthening its proteomics repertoire with a collaboration with Thermo Electron Corp. The two companies want to achieve greater integration between the Waters® ACQUITY UPLC™ system and Thermo’s full range of innovative mass spectrometers (including Thermo’s Finnigan™ TSQ Quantum™, LTQ™ Linear Ion Trap, LTQ FT™ Hybrid Linear Trap/Fourier Transform ICR and LTQ Orbitrap™ hybrid mass spectrometer). This integration will enable UPLC and mass spectrometry (MS) analyses to be combined more easily, and has the added benefit of increased throughput and performance. Results management software to handle the data performed using these techniques was also in demand.
“UPLC peaks are narrower and sharper, signal-to-noise ratios are higher, and run times are shorter – which are all key factors in enhancing the performance of virtually any mass spectrometer, thus bringing laboratories new levels of efficiency. For Waters customers, the seamless combination of UPLC technology with Thermo’s high-performance MS systems promises to bring laboratories more information per unit of time, resulting in greater productivity,” said Rohit Khanna, Vice President of Worldwide Marketing, Waters Corp.
Iain Mylchreest, Vice President and General Manager of Life Sciences Mass Spectrometry for Thermo, added, “This collaboration continues Thermo’s commitment to provide its customers with advanced technology options to improve overall system performance. Since mass spectrometers comprise one of Thermo’s largest and fastest growing product lines, and customers have seen the benefits of UPLC technology in combination with Thermo’s high performance mass spectrometry systems, it makes sense to give customers this front-end choice.”
Applied Biosystems completes acquisition of Ambion, Inc.’s Research Products Division
Ambion, Inc.’s Research Products Division, a leading provider of RNA analysis products, has been acquired by Applied Biosystems Group.
“We are excited about the many opportunities for sales growth and new products that we anticipate as a result of this strategic acquisition,” said Catherine M Burzik, President of Applied Biosystems. “We welcome Ambion’s employees and anticipate a smooth integration of product lines and personnel as we accelerate the delivery of more complete workflow solutions to our global customer base.”
Since it was founded in 1989, many of Ambion’s products for stabilizing, synthesizing, isolating, detecting, amplifying and quantifying RNA have become important tools in the genomics revolution. Examples of Ambion’s products include reagents associated with microRNA and RNA interference. These areas are of particular interest in the rapidly expanding study of the -omics. MicroRNA is involved in a number of processes, including early development, cell proliferation and differentiation, apoptosis, fat metabolism, and oncogenesis. RNA interference has uses as a biological mechanism of gene silencing.
Other Ambion products, such as small interfering RNA reagents, further add to the repertoire of RNA-related products that Ambion has produced to aid the burgeoning -omics arena.
Agilix Corp. proteomics labeling technology acquired by PerkinElmer
PerkinElmer’s position as a leader in quantitative proteomics has been strengthened by its acquisition of Agilix Corp.’s proprietary proteomics technology. Examples of the technology developed by Agilix include powerful molecule-labeling technology that uses isobaric mass tags to enable the simultaneous quantification of molecules, such as proteins, from multiple samples. This technology can also be used to enhance mass spectrometry, which is a common proteomic analysis method, thereby enabling the unambiguous detection and identification of proteins.
“The Agilix multiplex protein labeling technology represents a breakthrough in quantitative proteomics. This leading-edge technology reduces the process workflow in proteomics analysis, and enables more accurate studies in protein expression kinetics and pharmacodynamics,” said Neil Cook, Chief Scientific Officer, PerkinElmer Life and Analytical Sciences.
“PerkinElmer will use the intellectual property acquired from Agilix to develop a number of new technologies that will bring quantitative proteomics to a wider range of customers in drug discovery,” said Robert F Friel, President, PerkinElmer Life and Analytical Sciences.
GenoLogics and Proteome Software announce collaboration agreement
GenoLogics Life Sciences Software Inc. and Proteome Software have announced that they are to collaborate on the development of an integrated solution for proteomics research. The aim is to integrate GenoLogic’s ProteusLIMS™ laboratory and data management solution, and Proteome Software’s Scaffold software, thereby enabling advanced data visualization and mining of protein searches.
“The collaboration brings together the best of breed in scientific data management and visualization for protein search and analysis, and will benefit customers of both companies,” noted James DeGreef, Vice President Product Management for GenoLogics. “Researchers can take advantage of using multiple protein search engines in their clinical proteomics and biomarker research to gain more insight into the data and advance their research to the next level.”
Mark Pitman, Sales and Marketing Director for Proteome Software, said, “The synergy between GenoLogics and Proteome Software is such a natural one. We’re both committed to bringing proteomics researchers the cutting edge tools needed for the next generation of protein identification and analysis. We see great benefit for both companies’ customers coming out of this collaboration.”
‘Unraveling Collagen’ structure to be installed in Orange Memorial Park Sculpture Garden
On May 10th this year, a stainless steel sculpture of collagen is to be installed in the City of South San Francisco’s Orange Memorial Park Sculpture Garden. The creator of the piece, Julian Voss-Andreae, is a German-born artist with a background in quantum physics. This passion for science has inspired a series of sculptures that convey his deep sense of wonder and awe of life’s building blocks.
‘Unraveling Collagen’, as the name suggests, was inspired by the structure of collagen . Collagen is one of the most abundant proteins in the human body and provides structural support to the body, especially in bones, teeth, tendons, cartilage and ligaments. It was this flexible but tough quality that initially inspired Voss-Andreae.
Voss-Andreae uses scientific data that encode the shape of proteins as the starting point for his sculptures. Voss-Andreae chose to deviate from the precise molecular structure by opening the intertwining helices halfway up the structure, and molding the helices to create more fluid motion, similar to trees. “Suddenly the sculpture came to life and at the same time it turned into a metaphor for aging and growth,” noted Voss-Andreae.
Collagen is well known for its role in aging; it initially maintains the youthful elasticity of the skin, but as it degrades with age, the skin develops wrinkles. Besides this literal representation of aging as a physical process, the sculpture is also a metaphor of the wisdom and spiritual awakening that can come with old age.
The sculpture, which stands over 3.40 m, is one of a series of protein-based sculptures by Voss-Andreae. Other proteins in his collection include hemoglobin (Heart of Steel, 2005), green fluorescent protein (2004) and virus capsid protein (2003). Details of Voss-Andreae’s work can be found at www.julianvossandreae.com.
‘Unraveling Collagen’ will be on display at the Orange Memorial Park Sculpture Garden until spring 2008.