The complexity of protein involvement in organ function is poorly understood through the hyper- or hypo-expression of proteins.
It is well accepted that proteomic analysis is becoming more important to: investigate protein profiles of cells, biopsies and fluids; explore protein-based mechanisms of human disease; identify novel biomarkers for diagnosis, therapy and prognosis of multiple diseases; and discover new targets for drug development. Most of these foci are based on evidence from altered mapping of protein profiles under certain situations. Proteomic analysis was initiated to map the alterations of proteins, and then developed to correlate with protein and cell function, pathological changes, and medical prediction. It is now time to consider whether proteomic investigation can be linked and/or correlated with organ function and dysfunction. This is why a number of clinical medical journals have requested proteomic scientists and experts to make contributions regarding: molecular mechanisms of organ dysfunction; similarities and variations between etiologies; the identification of biomarkers for diagnosis and monitoring the process; the detection of therapeutic effects of new drugs; and follow-up the prognosis of patients with multiple-organ dysfunction syndrome (MODS) Citation[1].
Proteomic analysis for biomarkers has been extensively applied in a number of diseases (e.g., cancer, cardiovascular disease, acute renal allograft rejection, radiation and allergy). Protein expression data from proteomic studies have been correlated with clinical data such as histopathology, clinical functional measurements, medical imaging scores, patient demographics and clinical outcomes Citation[2]. Clinical proteomic studies are currently cataloged into three main models:
| • | To analyze altered patterns of protein contents in human blood, urine, secretary mucus and leaked fluid; | ||||
| • | To scan and map protein profiles of biopsies; | ||||
| • | To compare special variation between pathological changes and normal ones using laser-capture microdissection. | ||||
It is possible to measure protein profiles in highly selected cells from complex tissues and correlate them with disease-specific pathology; thereby, combining the morphological precision of microscopy with the power of molecular genetics, genomics and proteomics Citation[3]. More recently, imaging of tissue sections by mass spectrometry provides more detailed molecular figures containing information on both the abundance and distribution of many constituent compounds. Mass spectra are acquired directly from fresh frozen-tissue sections using matrix-assisted laser desorption/ionization mass spectrometry. It has been suggested that the availability of such molecular information helps clinicians improve therapeutic efficacy in the future Citation[4]. However, there are very few studies to determine the correlation between protein profiles and organ function, and between altered proteomics and organ dysfunction. It is even more difficult to explore the significance of proteomics in the development of multiple organ dysfunction, which is a systemic consequence of both acute and chronic diseases, and a critical and important phase of disease development Citation[5].
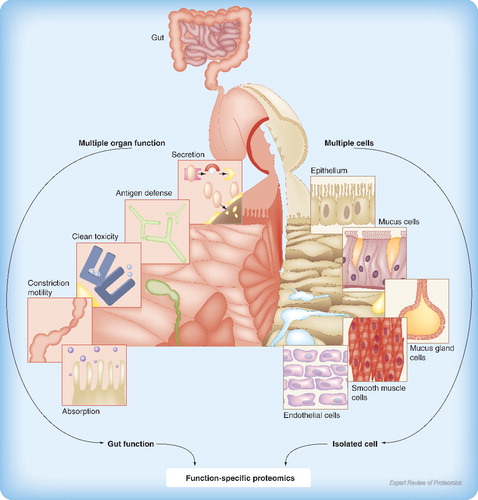
In general, organ dysfunction describes the development of biochemical and functional abnormalities in single or multiple organs Citation[5]. MODS is clinically considered as a sequential or concomitant occurrence of a significant derangement of function in two or more organ systems of the body, in the background of a critical illness. A more acceptable definition of MODS is ‘the presence of altered function in an acutely ill patient, such that homeostasis cannot be maintained without intervention’ Citation[6]. MODS has higher mortality and morbidity, and has more influencing factors in the pathogenesis; thus, it is more difficult for clinicians to investigate MODS than any single disease. For example, variations in proteomic profiles of epithelial cells exist based on location, disease types and phases, age, and organs, which may be one of many potential mechanisms responsible for the development of MODS Citation[7]. However, there are few studies to investigate whether proteomic profiles of cells can reflect or indicate the function of an entire organ. From systemic physiology, an organ has multiple functions and cell types; while a cell is involved in multiple functions, a function is performed by many cells. The complexity of protein involvement in organ function is poorly understood by the hyper- or hypo-expression of proteins. It is possible that one protein within multiple cell types is committed to only one organ function or many, while many proteins within one cell type may operate one function, as illustrated in .
It is encouraging that there is a growing number of proteomic scientists who are begining to explore the utilization of proteomics in the evaluation of cell or organ function. For example, a proteomic approach was employed to detect rat cardiac proteins that were differentially expressed or modified after exercise training Citation[8]. A total of 26 protein spot intensities were found to be significantly altered in hypertrophied hearts, with a marked increase in the expression of heat-shock protein 20. The question is whether or not these points are responsible for the formation of cardiac hypertrotrophy or involved in heart function. Kislinger and coworkers examined the protein composition of four subcellular compartments (cytosol, membrane-derived microsomes, mitochondria and nuclei) isolated by differential ultracentrifugation from healthy adult laboratory mouse brain, heart, kidney, liver, lung and embryonic placenta Citation[9]. The studies indicate that the proteomic scientists focused on using organ-selective gene products as biomarkers to monitor homeostatic perturbations associated with tissue-specific pathologies. Proteomic measurements have an advantage over mRNA profiling because they are able to deduce protein subcellular localization directly, thereby providing additional insight into the biological context of uncharacterized gene products that can lead naturally to testable hypotheses regarding function. It is the greatest challenge and opportunity to draw a clear and strong correlation between proteomic profiles and their associated organ function and/or dysfunction.
Special proteomic profiles may revolutionize proteomic sciences to clarify cell, tissue and organ function, and be a great contribution to understanding molecular mechanisms of organ function and/or dysfunction. The combination of proteomics with pathology is an example of investigating the potential involvement of proteins in the cell damage and morphometric alterations. It is definitely reasonable to believe that exploring the correlation of proteomics with organ function is the next step for both proteomic and clinical scientists, even though the mission seems impossible at present.
References
- Wang X, Adler KB, Chaudry IH, Ward PA. Better understanding of organ dysfunction requires proteomic involvement. J. Proteome Res.5, 1060–1062 (2006).
- Marko-Varga G, Lindberg H, Lofdahl CG et al. Discovery of biomarker candidates within disease by protein profiling: principles and concepts. J. Proteome Res.4, 1200–1212 (2005).
- Kiechle FL, Zhang X, Holland-Staley CA. The -omics era and its impact. Arch. Pathol. Lab. Med.128, 1337–1345 (2004).
- Chaurand P, Cornett DS, Caprioli RM. Molecular imaging of thin mammalian tissue sections by mass spectrometry. Curr. Opin. Biotechnol.17, 431–436 (2006).
- Wang XD. Organ dysfunction: a systemic consequence of acute and chronic diseases. J. Organ Dysfunction1, 2–3 (2005).
- Vincent JL. Organ dysfunction scores in critical illness. J. Organ Dysfunction1, 18–23 (2005).
- Zhao H, Adler KB, Bai C, Tang F, Wang XD. Epithelial proteomics in multiple organs and tissues: similarities and variations between cells, organs, and diseases. J. Proteome Res.5, 743–755 (2006).
- Boluyt MO, Brevick JL, Rogers DS, Randall MJ, Scalia AF, Li ZB. Changes in the rat heart proteome induced by exercise training: increased abundance of heat shock protein hsp20. Proteomics6, 3154–3169 (2006).
- Kislinger T, Cox B, Kannan A et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell125, 173–186 (2006).