Abstract
The slow down in the drug discovery pipeline is, in part, owing to a lack of structural and functional information available for new drug targets. Membrane proteins, the targets of well over 50% of marketed pharmaceuticals, present a particular challenge. As they are not naturally abundant, they must be produced recombinantly for the structural biology that is a prerequisite to structure-based drug design. Unfortunately, however, obtaining high yields of functional, recombinant membrane proteins remains a major bottleneck in contemporary bioscience. While repeated rounds of trial-and-error optimization have not (and cannot) reveal mechanistic details of the biology of recombinant protein production, examination of the host response has provided new insights. To this end, we published an early transcriptome analysis that identified genes implicated in high-yielding yeast cell factories, which has enabled the engineering of improved production strains. These advances offer hope that the bottleneck of membrane protein production can be relieved rationally.
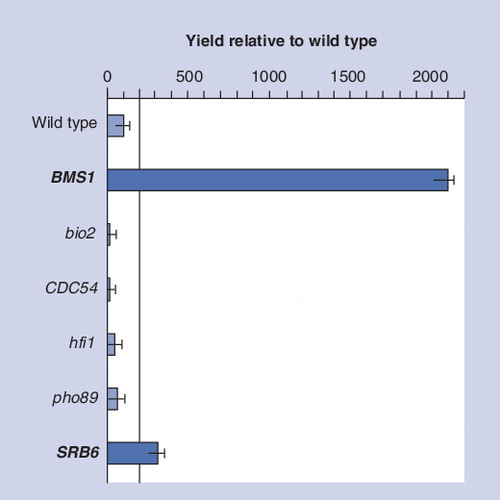
Fps1 yields, as assessed by immunoblot, are reported relative to wild type. Lower-case letters denote deletion strains, while upper-case letters denote strains with an upregulated expression of that particular gene. BMS1 and SRB6 are in bold as the yields obtained for these strains in a production screen exceeded the threshold (more than twice that of wild type) required for further study.
Bottleneck of recombinant membrane protein production
Understanding membrane proteins is vital for the development of new drugs in the fight against human disease Citation[1]. As they are not highly abundant naturally, membrane proteins must be produced recombinantly for the detailed studies that will reveal their biochemical, functional and structural characteristics. Despite this need, obtaining high yields of functional, recombinant membrane protein remains a major bottleneck in contemporary bioscience Citation[2]. We have shown that the root of the problem is understanding the host organism Citation[3] and, in particular, its response to the production of recombinant proteins in its cell membranes. Our rational approach to this problem is in stark contrast to most protein production strategies that rely on repeated rounds of trial-and-error optimization and cannot provide a mechanistic insight, which is also true of approaches that rely on the mutation of the protein target to improve its production yields Citation[4]. The use of postgenomic array methods have been key to this rational approach to optimization Citation[3] in yeast Citation[5]; in one such example, we reported 39 host cell genes whose expression was significantly altered when high-yielding production conditions were compared with low-yielding conditions in Saccharomyces cerevisiae. Although similar studies have subsequently been performed in other hosts Citation[6,7], mechanistic insight into successful recombinant protein production has remained elusive Citation[8]. Building on our comparative transcriptome analysis Citation[3], we recently showed that deletion of three S. cerevisiae genes could each increase protein yields. In particular, overexpression of a fourth gene from our list, BMS1, could be specifically tuned to maximize yields of a range of membrane proteins Citation[9]. By altering the amount of BMS1 transcript, the metabolism of high-yielding cultures was changed substantially and coincided with the ratio of ribosomal subunits being perturbed, offering, for the first time, an insight into the actual mechanisms involved.
Designing appropriate experiments to address the bottleneck
The predominant technique for obtaining a description of a global cellular response to a genetic or environmental change is to use DNA microarrays. Modern arrays comprise amplified cDNA or oligonucleotides (typically ranging from 25 to 60 nucleotides in length), which are either spotted or directly synthesized onto a solid support in various layouts. Widely used microarray chips are marketed by Agilent Technologies Incorporated Citation[101] and Affymetrix Incorporated Citation[102], although other platforms, as well as home-made options, exist. An article from 2006 in Expert Review of Molecular Diagnostics, provides useful pointers in the selection of appropriate microarray platforms and the principles of experimental design Citation[10].
We previously used nylon filter arrays generated in Louise Showe’s laboratory Citation[103] in order to understand the yeast response to stresses, such as slow glucose uptake rates Citation[11] and membrane protein production Citation[3]. In each case, our aim has been to achieve a list of all genes that were potentially correlated with these phenotypes, but which was not too large to be subsequently validated by independent analysis, such as quantative (Q)-PCR or strain engineering.
Typically, a pair-wise comparison of RNA preparations in two to six biological repeats is performed for a reference and several specific test conditions. Obtaining a manageable number of genes for validation and testing that are correlated with significantly changed mRNA levels requires a robust experimental design. This can be achieved by sampling cells growing at the same growth rate but over a range of optical densities (ODs; e.g., 0.5, 1.0 and 1.5) rather than multiple sampling at a single OD. In addition, introducing further ‘redundancy’ into the test conditions can filter out nonspecific effects. For example, we examined two different growth conditions that both led to relatively low protein yields compared with normal conditions, and looked for changes in mRNAs that occurred in the same direction in both sets. This highlighted 39 genes. It is notable that other array studies of S. cerevisiae, albeit not focused on protein production, typically present many more genes as being significantly regulated. For example, a shift from 27 to 37°C for 30–80 min yielded 447 genes Citation[12], while there were 150 upregulated genes following a shift to pH 7.4 Citation[13]. A shift from 30 to 10°C for 8 h induced a change in 1024 genes Citation[14].
The 39 genes that we found were validated against the list obtained when comparing the single growth condition that led to higher protein yields compared with normal conditions. shows an example of the data obtained. In all cases, genes that were downregulated under low-yielding conditions were upregulated under high-yielding conditions and vice versa. This lent confidence that these genes had a role in high-yielding protein production, which was subsequently further validated by strain engineering . Specifically, we used this approach to characterize the role of BMS1Citation[9] and to engineer the first yeast production strain (GB0813253.2) by rational design Citation[8].
In another study, we examined the ability of a respiratory yeast strain, V5.TM6*P, to grow on glucose compared to its respiro-fermentative parent, V5. We were able to improve the analysis even further by comparing all previous analyses of the yeast diauxic shift, which had been first studied by microarray in 1997 Citation[15]. Again, we compared the two strains (n = 12) over a range of sampling points (36–5 g/l glucose), rather than taking multiple samples at a single point Citation[11]. Our dataset gives a remarkably complete view of the involvement of genes in the tricarboxylic acid cycle, glyoxylate cycle and respiratory chain in the expression of the phenotype of V5.TM6*P. Furthermore, 88% of the transcriptional response of the induced genes in our dataset could be related to the potential activities of just three proteins: Hap4, Cat8 and Mig1. Overall, the data supported genetic remodeling in V5.TM6*P consistent with a respiratory metabolism that is insensitive to external glucose concentrations.
Validation by Q-PCR & subsequent strain engineering
Validation of array results by Q-PCR or strain engineering is an essential next step. Our protein production strain, in which the ribosomal gene BMS1 was upregulated by a factor of 6–7 compared with wild-type, gave corresponding yield improvements of a factor of 2–70 for a range of membrane proteins Citation[9]. This rationally engineered strain complements strains resulting from previous speculative deletion strategies. In S. cerevisiae, for example, deletion of two vacuolar proteases increased yields of the G-protein-coupled receptors, Ste2, by a factor of 10 Citation[16], while in Pichia pastoris strain SMD1163, the deletion of two protease genes (PEP4 and PBR) boosted membrane protein production Citation[17]. Minimizing the unfolded protein response (UPR) of S. cerevisiae has also improved the yields of certain functional membrane protein Citation[18] that induce the UPR.
Expert commentary
The production of recombinant membrane proteins is finally being recognized as a science rather than an art. Using systems biotechnology, it is now possible to identify changes in gene expression in the host cell that are associated with high yields of these potential drug targets. We have used the results of one such study to engineer high-yielding yeast strains and are now in a position to gain further mechanistic insights using postgenomic techniques.
Five-year view
We have shown that comparative transcriptome analysis of S. cerevisiae host cells during high- versus low-yielding recombinant protein production experiments can guide strain engineering. In particular, by titrating the overexpression of BMS1, a ribosome biogenesis gene, the functional yields of a range of membrane proteins can be improved by a factor of 2–70. This discovery is the basis of a recent patent application (GB0813253.2), which also describes three additional engineered strains. With the very recent publication of a curated P. pastoris genome Citation[19], we will now be able to transfer this technology to a second industrially relevant yeast species and we further predict that this approach will be used for a range of host cells and protein targets. As array techniques have improved, it is now possible to relate changes in mRNA with changes at the protein level. A comparative proteome analysis of membrane versus soluble protein production in E. coli highlighted that the cytoplasmic membrane protein translocation machinery might be limiting in low-yielding bacterial transformants Citation[6]. Interestingly, we found no evidence for this in yeast Citation[9], which highlights clear differences between prokaryotic and eukaryotic cells. With time, the ability to combine the outputs from these types of studies with those from metabolomic approaches will contribute to a true systems biotechnological description of recombinant membrane protein production in yeast.
Table 1. Array data from a transcriptome analysis of yeast strains giving high or low yields of recombinant membrane protein.
Key issues
• There is a move toward structure-based drug design in modern discovery pipelines.
• Membrane proteins, especially G-protein-coupled receptors and ion channels, are the most important class of new drug targets.
• The recombinant production of membrane proteins is a challenging task.
• Array-based techniques are finally shedding light on the mechanisms underpinning high yields of recombinant proteins in yeast cells.
• Careful experimental design is critical to identifying genes whose expression is correlated with highly recombinant protein yields.
• These experiments are an efficient route to engineering new, high-yielding production strains.
Financial & competing interests disclosure
The authors are supported by EPSRC CASE awards with Alpha Biologics Ltd and Applikon Biotechnology Ltd, a BBSRC CASE award with AstraZeneca Ltd and a Technology Strategy Board grant with Glycoform Ltd and Polytherics Ltd, as well as European Commission contracts LSHG-CT-2004–504601 (E-MeP), LSHG-CT-2006–037793 (OptiCryst) and Grant Agreement Number 201924 (EDICT) to RMB. The BBSRC supports bioreactors and a flow microcalorimeter in the RMB laboratory through an REI award. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Schlyer S, Horuk R. I want a new drug: G-protein-coupled receptors in drug development. Drug Discov. Today11(11–12), 481–493 (2006).
- Forstner M, Leder L, Mayr LM. Optimization of protein expression systems for modern drug discovery. Expert Rev. Proteomics4(1), 67–78 (2007).
- Bonander N, Hedfalk K, Larsson C et al. Design of improved membrane protein production experiments: quantitation of the host response. Protein Science14(7), 1729–1740 (2005).
- Martinez Molina D, Cornvik T, Eshaghi S et al. Engineering membrane protein overproduction in Escherichia coli. Protein Sci.17(4), 673–680 (2008).
- Bill RM. Yeast – a panacea for the structure-function analysis of membrane proteins? Curr. Genet.40(3), 157–171 (2001).
- Wagner S, Baars L, Ytterberg AJ et al. Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteomics6(9), 1527–1550 (2007).
- Wagner S, Klepsch MM, Schlegel S et al. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl Acad. Sci. USA105(38), 14371–14376 (2008).
- Sevastsyanovich Y, Alfasi S, Cole J. Recombinant protein production: a comparative view on host physiology. New Biotechnol.25(4), 175–180 (2009).
- Bonander N, Darby RA, Grgic L et al. Altering the ribosomal subunit ratio in yeast maximizes recombinant protein yield. Microb. Cell. Fact.8, 10 (2009).
- Ahmed FE. Microarray RNA transcriptional profiling: part I. Platforms, experimental design and standardization. Expert Rev. Mol. Diagn.6(4), 535–550 (2006).
- Bonander N, Ferndahl C, Mostad P et al. Transcriptome analysis of a respiratory Saccharomyces cerevisiae strain suggests the expression of its phenotype is glucose insensitive and predominantly controlled by Hap4, Cat8 and Mig1. BMC Genomics9, 365 (2008).
- Gasch AP, Spellman PT, Kao CM et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell.11(12), 4241–4257 (2000).
- Serrano R, Ruiz A, Bernal D, Chambers JR, Arino J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol.46(5), 1319–1333 (2002).
- Sahara T, Goda T, Ohgiya S. Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. J. Biol. Chem.277(51), 50015–50021 (2002).
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science278(5338), 680–686 (1997).
- David NE, Gee M, Andersen B et al. Expression and purification of the Saccharomyces cerevisiae α-factor receptor (Ste2p), a 7-transmembrane-segment G protein-coupled receptor. J. Biol. Chem.272(24), 15553–15561 (1997).
- Andre N, Cherouati N, Prual C et al. Enhancing functional production of G protein-coupled receptors in Pichia pastoris to levels required for structural studies via a single expression screen. Protein Sci.15(5), 1115–1126 (2006).
- Griffith DA, Delipala C, Leadsham J, Jarvis SM, Oesterhelt D. A novel yeast expression system for the overproduction of quality-controlled membrane proteins. FEBS Lett.553(1–2), 45–50 (2003).
- De Schutter K, Lin YC, Tiels P et al. Genome sequence of the recombinant protein production host Pichia pastoris. Nature Biotechnol.27(6), 561–566 (2009).
Websites
- Agilent Technologies Incorporated www.agilent.com
- Affymetrix Incorporated www.affymetrix.com
- Professor Louise Showe’s laboratory http://showelab.wistar.upenn.edu/Wistar_Showe_Lab_Gene_Lists.htm