ABSTRACT
Some of the world’s highest-altitude butterflies belong to the tribe Pierini of the family Pieridae. Two nominal species of Baltia occur in arid-semiarid oreal environments in Central Asia, the Himalaya, and Pamir to over 5500 m. At least 13 species currently placed in four genera (Phulia, Infraphulia, Pierphulia, Piercolias) occur in similar environments at similar altitudes in the high Andes. These genera all share numerous morphospecializations whose functional relation to the oreal environment is not understood. Their evolutionary and biogeographic relationships have been debated for over a century. We performed a phylogenetic analysis based on sequencing portions of the mitochondrial cytochrome oxidase subunit I and subunit II regions (COI and COII), incorporating all the genera but Piercolias and a variety of suspected relatives. The results from analyses of COI and COII were compared to relationships inferred from morphological and ecological characters. We conclude that Baltia is not the sister-group of the Andean genera, which are clearly nested within a Neotropical clade. The “Camelid scenario” deriving all the genera from a common ancestor no longer appears viable.
Introduction
The geological ages of mountain ranges should constrain the time frame in which their endemic high-altitude biota could have evolved. The exigencies of life in the oreal zone apparently select for characteristic phenotypic (morphological) syndromes—the giant rosette-shrub growth form of tropical oreal plants is a dramatic example—although the specific functions of the component traits may be poorly if at all understood (CitationDescimon, 1986). This fact predisposes these organisms to convergent evolution, which may be a serious impediment to understanding their phylogeny and historical biogeography. Remarkably little attention has been paid to the question of how quickly such syndromes can arise, despite the availability of molecular tools adapted to quantifying genetic distance and reconstructing phylogeny.
Some of the world's highest-altitude butterflies belong to the tribe Pierini of the family Pieridae, reaching 5500 m or higher in the Andes (S. Halloy, personal communication), Himalaya (CitationMani, 1986), Pamir and other high Central and South Asian ranges. All of these ranges, and the landscapes and semiarid to arid biotopes in which the butterflies are endemic, are believed to be quite young—no older than late Tertiary and most likely Quaternary (Andes: CitationLamb et al., 1997; CitationClapperton, 1993; Asia: CitationKalvoda, 1992; CitationShackleton and Chengfa, 1988). By most accounts the oreal climates inhabited by these butterflies are consequent to their tectonic evolution (CitationRaymo and Ruddiman, 1992).
The true relationship between the South American and Asian species has been debated vigorously for well over a century. There are two broad historical-biogeographic scenarios for them in the literature: either the Andean and Asian genera arose independently in situ within the constraints of their respective tectonic time-lines, or they arose from a common ancestor farther in the past, arriving at their current distributions by long-distance dispersal. The latter has been compared (CitationShapiro, 1992) to the well-known “Camelid scenario” of CitationMatthew (1915). The camelids apparently originated in North America, dispersed to both Asia and South America, and subsequently became extinct on their continent of origin, giving rise to an Andean-Asian disjunct distribution. In the former case the closest relatives of each should be found in its own geographic region. In the latter, the Asian oreal taxa should be most closely related to the Andean ones. We bring molecular and phylogenetic methods to bear in hopes of resolving the issue.
The butterfly genera are Baltia in Asia (two nominal species) and Phulia, Infraphulia, Pierphulia, and Piercolias (13 nominal species recognized in the last revision: CitationField and Herrera, 1977) in the Andes. (In older literature Infraphulia and Pierphulia were lumped into Phulia, and Piercolias is usually called Andina. Our unpublished studies indicate that cryptic species may exist on both continents.) The enigmatic, monotypic Reliquia, from the oreal of the Sierra Nevada de Santa Marta, Colombia (CitationAckery, 1975) may or may not be related.
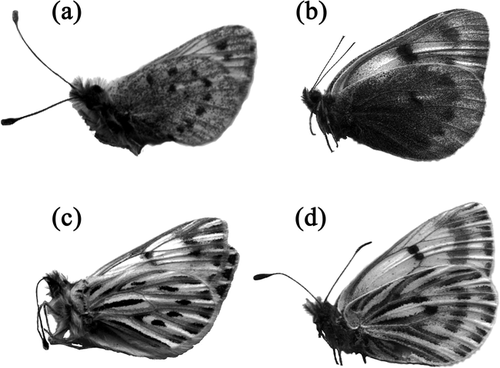
One of the two species of Baltia, B. butleri, is superficially extremely similar to Phulia and Infraphulia (). The other, B. shawi, is equally similar to Pierphulia and Piercolias (). These resemblances have occasioned much pre-cladistic and pre-molecular phylogenetic speculation. Most of the resemblances can be at least conceived as functionally related to the oreal environment (wing shape, venation, and pattern are all probably significant in aerodynamics and thermoregulation). The anatomy of the genitalia has been interpreted as sheltered from such selective influences, hence unlikely to be convergent, and useful as an indicator of true relationships (CitationKlots, 1932). Close similarities have been alleged in the genitalia of Baltia and the Andean genera, leading CitationField (1958) and CitationField and Herrera (1977) to argue for a phylogenetic relationship. The female genitalia of Baltia and Piercolias, which are almost completely internal, are described as nearly identical.
Figure 1 Representative specimens of four taxa discussed in the text: (a) Pierphulia rosea, Chile; (b) Baltia shawi, Afghanistan; (c) Phulia nymphula, Chile; (d) Baltia butleri, Tibet. The two morphologically convergent pairs are P. rosea / B. shawi, and P. nymphula / B. butleri.
The historical debate was forged within a tradition of attributing the high-Andean biota to invasion from the Northern Hemisphere (CitationShapiro, 1992). A variety of evolutionary hypotheses and putative ancestors, as noted above, are posited by CitationDixey (1894), CitationElwes (1895), CitationGrote (1900), CitationRoeber (1908–1924), CitationKlots (1932), CitationForster (1958), CitationField (1958), CitationMani (1968, Citation1986), CitationField and Herrera (1977), CitationDescimon (1986), and CitationShapiro (1992).
There is a fossil, Miopieris talboti from the Upper Miocene of Swabia, southwest Germany (CitationZeuner, 1942), which appears related to some or all of the genera of interest, but lacks most relevant characters except the wing venation and is not useful in resolving relationships. We thus must rely on what can be learned from extant taxa. If molecular phylogenies are more informative in general than those derived from characters subject to convergence, a molecular vs. morphological study of these genera should be helpful. If the “Camelid scenario” is valid, phylogenies based on both types of data should agree. If the Andean and Asian animals are convergent, the two phylogenies should disagree.
Materials and Methods
We did not seek to generate a broad phylogeny of the Pierini. Rather, we used the literature to select genera that had been suggested as relatives of the oreal pierines: Aporia in the Old World, Pontia (Synchloe) throughout the Holarctic, the Andean-Patagonian Tatochila and Hypsochila, and the Neotropical Ascia. CitationField and Herrera (1977) considered the Andean oreal genera as specialized derivatives of Tatochila, with Hypsochila intermediate. Several authors postulated Baltia as a specialized derivative of Pontia; some suggested Pontia as the ancestor of Tatochila and thus, indirectly, of the Andean oreal genera. For outgroups we chose Colias eurytheme, as well as the bizarre, taxonomically isolated Eucheira socialis, a Mexican montane endemic suspected of being an ancient relict.
Twenty-two recent specimens representing 15 species were obtained and sequenced (see for collection and locality information); a C. eurytheme sequence was taken from GenBank (Accession #AF044024). We were unable to obtain recent material of Piercolias or Reliquia, and they are excluded. Except for a few mounted museum specimens, most of the material had been stored at −20°C until use. DNA extractions were performed with the Purgene DNA isolation kit (Gentra Systems, Minneapolis, MN, U.S.A.), and precipitated DNA was dried and resuspended in 200 µL of H20. The basal parts of abdomens were extracted; wings, legs, and genitalia of all specimens were retained as vouchers and will be deposited in the Bohart Museum of Entomology, University of California (UC), Davis.
Table 1 Locality and collection information for specimens studied. Numbers in parentheses indicate the number of individuals sampled of each species. Collectors as follows: all specimens from Chile, California, and Texas (A. M. Shapiro); A. crataegi (M. Jagelka); B. butleri (M. Zubrik); B. shawii (R. Westphal); E. socialis (D. Underwood). One Colias eurytheme (not listed) sequence was obtained from GenBank.
The following primers were used in polymerase chain reactions (PCR): to amplify a portion of the mitochondrial cytochrome oxidase subunit I region (COI): k698 (5′ TAC AAT TTA TCG CCT AAA CTT CAG CC) and k525 (5′ ACT GTA AAT ATA TGA TGA GCT CA); and for a portion of the mitochondrial cytochrome oxidase subunit II region (COII): EVA (5′ GAG ACC ATT ACT TGC TTT CAG TCA TCT 3′) and PATRICK (5′ CTA ATA TGG CAG ATT ATA TGT ATT GGA 3′) (CitationCaterino and Sperling, 1999). PCR amplifications were run according to the following protocol: (1) 94°C for 2 min; (2) 39 cycles of 94°C for 1 min, 48°C for 45 s, and 72°C for 1 min 45 s; (3) 72°C for 10 min. PCR products were sequenced in both directions on ABI 3730 (COI) and ABI 377 (COII) automatic sequencers at the DNA Sequencing Facility, Division of Biological Sciences, UC Davis, producing 612 bp of COI and 501 bp of COII that were consistently readable. The SeqEd computer program (v.1.0.3, Applied Biosystems, Foster City, CA, U.S.A.) was used to aid in the visual alignment of sequences.
Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian Maximum Likelihood (BML) were used with the COI and COII sequences to examine the relationships among taxa. Using hierarchical likelihood ratio tests (α < 0.01) in the program Modeltest v.3 (CitationPosada and Crandall, 1988), 56 models of evolution were evaluated for the portions of COI and COII sequenced ().
Table 2 DNA substitution models selected using maximum likelihood ratio tests.
MP (1000 bootstrap pseudoreplicates) and ML (100 pseudoreplicates) analyses were done with the program PAUP* v.4.0b10 (CitationSwofford, 2002). Bayesian analysis was performed with the program MRBAYES (CitationHuelsenbeck and Ronquist, 2001; available online at http://morphbank.ebc.uu.se/mrbayes/), which uses a metropolis-coupled, Markov-chain Monte Carlo method to estimate posterior probabilities associated with nodes. A total of 2 million pseudoreplicates were run in MRBAYES; trees were sampled every 100 generations. A “burn-in” value was estimated by plotting log-likelihood values against generation time, and a consensus tree with posterior probabilities was then generated in PAUP* for all sampled trees posterior to the “burn-in” generation.
For MP and ML analyses, the portions of COI and COII sequenced for each individual were analyzed as contiguous sequences following evaluation with a partition-homogeneity test in PAUP* (1000 replicates), which returned P-values near or below 0.05, suggesting acceptable congruence of the phylogenetic information in each region (one run of the test produced P = 0.053, and a second run produced P = 0.042). Bayesian analyses were implemented with six partitions, such that each codon position of each gene was modeled separately (see for the models used for each data partition).
A morphological matrix () was generated for 13 taxa using data already available from published descriptions or our own work. Characters were derived from both adults and immatures. Host plant, which is often phylogenetically informative in butterflies (CitationBraby et al., 2006), was also used as a character. A number of potentially useful characters were omitted because relevant information was lacking or difficult to interpret for some of the genera. The data were used in the computer program PAUP* to identify the most parsimonious relationships among taxa. The Shimodaira-Hasegawa (SH) Test was then used to compare the topology of the trees generated with parsimony analysis of morphological and ecological characters to the best maximum-likelihood sequence tree (combined COI and COII), with similarity rejected at the level p < 0.05 (1000 RELL bootstrap pseudoreplicates were used).
Table 3 Morphological character matrix. Characters and codes are as follows: (1) number of radial veins: 0 = 2, 1 = 3, 2 = 4; (2) size of pulvilli: 0 = large, 1 = reduced; (3) size of paronychia: 0 = large, 1 = reduced; (4) M1 stalked on forewing: 0 = no, 1 = yes; (5) M2 stalked on forewing: 0 = no, 1 = yes; (6) length of third segment of pulpus: 0 = very short, 1 = short, 2 = “normal”; (7) tegumen size: 0 = very short, 1 = large, 2 = “normal”; (8) apex of forewing rounded: 0 = rounded, 1 = angled; (9) larvae gregarious: 0 = no, 1 = yes; (10) conical development of lower juxta: 0 = not, 1 = somewhat, 2 = pronounced; (11) tibial spurs on middle and hind legs: 0 = no, 1 = yes; (12) harpe simple, rounded: 0 = no, 1 = yes; (13) overall VHW “thermal” melanization: 0 = no, 1 = yes; (14) wing vein darkening: 0 = veins not melanized relative to ground color, 1 = thin black line directly on veins, 2 = dark edging on veins, veins not themselves darkened, 3 = broad dark lines including the veins; (15) chevrons concolorous with vein-lines on ventral hindwing: 0 = no chevrons, 1 = no, 2 = yes (Hypsochilla variable); (16) direction of “arrow” of ventral hindwing chevrons: 0 = no chevrons, 1 = point marginad, 2 = no point, 3 = point basad; (17) pupal attachment: 0 = pendant, 1 = appressed; (18) pupal angularity: 0 = lightly angular, 1 = heavily angular, 2 = obtect; (19) larval host: 0 = woody, non-legume, non-Brassicaceae, 1 = Brassicaceae, 2 = legumes; (20) R3 and R4+5 very stalked, veins themselves short: 0 = no, 1 = yes.
Results and Discussion
Analysis of the portions of COI and COII sequenced produced 187 variable sites with 140 informative sites from COI, and 132 variable sites with 103 informative sites from COII. Sequences from all specimens are available on GenBank (Accession nos. DQ463374 to DQ463417). Saturation plots examined for each codon position in both COI and COII gave no indication that any of the sites were saturated with multiple substitutions.
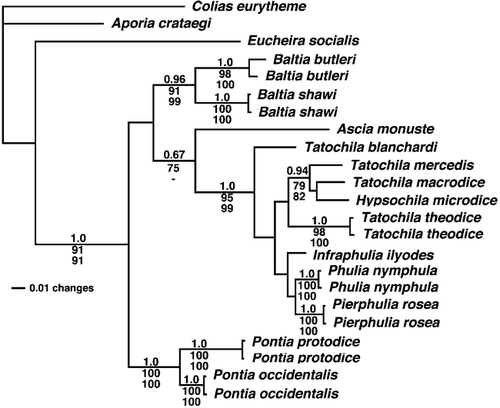
The ML, BML, and MP phylogenies put the Andean oreal genera into a monophyletic group with Andean non-oreal genera (as predicted by CitationField and Herrera, 1977), but group no Andean genus with the Northern Hemisphere (). Baltia is also monophyletic in these phylogenies, as is Pontia (Synchloe). The relationships described in falsify the hypothesis, which CitationField (1958) had posited, that Baltia and the Andean oreal genera are sister-groups.
Figure 2 Phylogenetic tree resulting from maximum likelihood analysis. Bayesian maximum likelihood analysis produced a tree with identical topology. The three values shown at nodes are: (1) Bayesian posterior probabilities (listed on top); (2) percent recovery in maximum likelihood bootstrap analysis (listed in the middle); and (3) percent recovery in maximum parsimony bootstrap analysis (listed below). Support is only shown for nodes with one of the following: posterior probabilities greater than 0.95 or bootstrap values equal to or greater than 70. A hyphen (-) indicates nodes that were not resolved in parsimony analysis. Branches are scaled to the number of substitutions per site. See for models used in analyses. Models simpler than those shown in (i.e., Jukes-Cantor and Kimura 2-parameter) gave qualitatively identical results (reciprocal monophyly of the focal lineages in the Old and New Worlds).
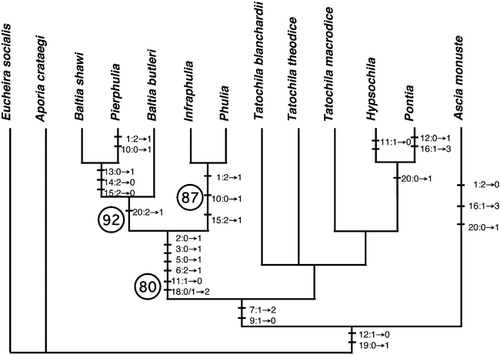
Nine equally parsimonious trees were produced by analyses of morphological and ecological characters. The topologies of these trees are similar in that a clade composed of Baltia shawi, Baltia butleri, Phulia, Infraphulia, and Pierphulia was recovered with 80% bootstrap support. An exemplar of the nine most parsimonious trees is shown in (the clade comprised of the five focal taxa is also present in the consensus tree, not shown). The results from parsimony analyses of morphological and ecological data unsurprisingly reflect the historic ambiguity found in narrative phylogenies—imbedding both Baltia species among Andean taxa but not as each other's sister-species. These results underscore that, at least at the coarse level of resolution used to date in scoring morphological characters, convergence and possibly reversals run rampant in this group. The nine equally parsimonious trees are all statistically different from the combined ML COI and COII tree at the level of P < 0.0001. If we place greater confidence in molecules than morphology, both sets of morphologically similar taxa (B. shawi / P. rosea; B. butleri / P. nymphula, I. ilyodes) are convergent.
Figure 3 One of nine equally parsimonious trees derived from analysis of morphological characters (only unambiguous state changes are shown). Numbers coding characters and character states are presented in . Circled numbers indicate percent recovery in bootstrap analysis (1000 pseudoreplicates); numbers shown are the only bootstraps greater than 50.
The internal structure of the Andean clade is complicated by problems with the existing taxonomy. CitationField and Herrera (1977), as earlier noted, saw Hypsochila as an intermediate step in the evolution of the oreal genera from a generalized, lowland Tatochila ancestor. At that time little was known about these butterflies except their adult morphology. Subsequent studies of the immature stages, biology, and host plants have suggested that both Tatochila and Hypsochila are almost certainly polyphyletic (CitationShapiro, 1991 and unpublished). The species we sequenced were selected to represent the perceived austral diversity in Tatochila. T. macrodice (central Andes, Puna, and Altiplano) and T. mercedis (lowland Mediterranean Chile) are fully interfertile and biologically conspecific, part of a large polytypic complex ranging from Colombia to Tierra del Fuego (CitationShapiro, 1991), but they did not fall out as sister-taxa here. Hypsochila microdice fell squarely within this cluster, despite the fact that on morphological grounds it belongs to a different genus! Tatochila theodice belongs in its own monotypic genus, based on both adult and early-stage characters. Whatever the proper relationships at this level and the implied insensitivity of the COI and COII sequences to them, they do not affect the higher-level question this study addresses. The position of Ascia, as recovered by ML, suggests that the entire Andean group of genera may be derived from the lowland tropics rather than from Holarctic ancestors, as commonly supposed (however, MP and BML failed to support this node).
Independently of our efforts, CitationBraby et al. (2006), in a sweeping phylogenetic survey of the Pieridae involving some of the same taxa but different molecular markers, find that Baltia (they had butleri only) clusters with Pontia and the Andean oreal Phulia, Pierphulia, and Infraphulia cluster with Tatochila, Hypsochila, and Theochila (which we did not have). Like us, they had neither Reliquia, which is morphologically very similar to Pontia, nor Piercolias. Despite differences in detail, largely due to having used several different taxa, their results agree with ours in rejecting the “Camelid scenario” for the oreal genera.
Despite persistent ambiguities in the overall phylogeny, we conclude that Baltia and the Andean oreal genera are not sister-taxa and do not form a clade antedating the current geographic pattern, as in the Camelidae. Their remarkable similarities are convergent (extending even to the genitalia), and must be accommodated within the Quaternary time frame of their oreal biomes—suggesting quite rapid morphological evolution and, in the Andes, taxonomic differentiation.
Acknowledgments
Collection of material from the Andes was supported by NSF grant BSR-83-06922 and two travel grants from the Committee on Research of the Academic Senate, UC Davis, all to Shapiro. The Committee on Research also supported sequencing. We thank Brad Shaffer for the use of his facilities, Tom Near for help with the analyses, and Michael Braby and Naomi Pierce for sharing their unpublished results with us.
References Cited
- Ackery, P. R. 1975. A new pierine genus and species with notes on the genus Tatochila (Lepidoptera: Pieridae). Bulletin of the Allyn Museum 30:1–9.
- Braby, M. F. , R. Vila , and N. E. Pierce . 2006. Molecular phylogeny and systematics of the Pieridae (Lepidoptera: Papilionoidea): higher classification and biogeography. Zoological Journal of the Linnaean Society 147:238–275.417.
- Caterino, M. S. and F. A. H. Sperling . 1999. Papilio phylogeny based on mitochondrial cytochrome oxidase I and II genes. Phylogenetics and Evolution 11:122–137.
- Clapperton, C. M. 1993. Quaternary Geology and Geomorphology of South America Amsterdam Elsevier. 779.
- Descimon, H. 1986. Origins of lepidopteran faunas in the high tropical Andes. In: Vuilleumier, F. and M. Monasterio , editors. eds. High-altitude Tropical Biogeography New York Oxford. 500–532.
- Dixey, F. A. 1894. On the phylogeny of the Pierinae, as illustrated by their wing-markings and geographical distribution. Transactions of the Entomological Society of London 1894:2 249–335.
- Elwes, H. J. 1895. Geographical distribution of butterflies. Transactions of the Entomological Society of London 1894:6 lii–1xxxiv.
- Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution 17:368–376.
- Field, W. D. 1958. A redefinition of the butterfly genera Tatochila, Phulia, Piercolias , and Baltia , with descriptions of related genera and subgenera. Proceedings of the United States National Museum 108:103–131.
- Field, W. E. and J. Herrera . 1977. The pierid butterflies of the genera Hypsochila Ureta, Phulia Herrich-Schaffer, Infraphulia Field, Pierphulia Field, and Piercolias Staudinger. Smithsonian Contributions to Zoology 232:1–64.
- Forster, W. 1958. Die Tiergeographischen Verhaktnisse Boliviens. In: Becker, E. C. , editor. ed. Proceedings of the Tenth International Congress of Entomology (1956) Ottawa Mortimer. vol. 1:843–846.
- Grote, A. R. 1900. The descent of the pierids. Proceedings of the American Philosophical Society 39:4–67.
- Hasegawa, M. , K. Kishino , and T. Yano . 1985. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution 22:160–174.
- Huelsenbeck, J. P. and F. Ronquist . 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754–755.
- Kalvoda, J. 1992. Geomorphological record of the Quaternary orogeny in the Himalaya and the Karakoram Amsterdam Elsevier. 316.
- Klots, A. B. 1932. A generic revision of the Pieridae (Lepidoptera), together with a study of the male genitalia. Entomologica Americana 12:139–204.
- Lamb, S. , L. Hoke , L. Kennan , and J. Dewey . 1997. Cenozoic evolution of the Central Andes in Bolivia and northern Chile. In: Burg, J. P. and M. Ford , editors. eds. Orogeny through Time Special Publication 121. London The Geological Society. 237–264.
- Mani, M. S. 1968. Ecology and Biogeography of High-Altitude Insects The Hague Junk. 527.
- Mani, M. S. 1986. Butterflies of the Himalaya The Hague Junk. 181.
- Matthew, W. D. 1915. Climate and evolution. Annals of the New York Academy of Sciences 24:171–318.
- Posada, D. and K. A. Crandall . 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818.
- Raymo, M. E. and W. F. Ruddiman . 1992. Tectonic forcing of late Cenozoic climate. Nature 359:117–122.
- Roeber, J. 1908–1924. (South American Pieridae). In: Seitz, A. , editor. ed. The Macrolepidoptera of the World Stuttgart Kernen. 5:53–55–56.97–98.1015. 1016. 1024; plates 18, 28, 192, 194.
- Shackleton, R. M. and C. Chengfa . 1988. Cenozoic uplift and deformation of the Tibetan Plateau: the geomorphological evidence. Philosophical Transactions of the Royal Society of London, (A) 327:365–377.
- Shapiro, A. M. 1991. The zoogeography and systematics of the Argentine Andean and Patagonian pierid fauna. Journal of Research on the Lepidoptera 28:137–238.
- Shapiro, A. M. 1992. Why are there so few butterflies in the High Andes?. Journal of Research on the Lepidoptera 31:35–56.
- Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b.10. Sunderland, MA Sinauer.
- Tamura, K. and M. Nei . 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512–526.
- Zeuner, F. E. 1942. Two new fossil butterflies of the family Pieridae. Annals and Magazine of Natural History 9:409–416.