Abstract
Follistatin has been reported as a candidate gene for polycystic ovarian syndrome (PCOS) based on linkage and association studies. In this study, investigation of polymorphisms in the FST gene was done to determine if genetic variation is associated with susceptibility to PCOS. The nucleotide sequence of human follistatin and the protein sequence of human follistatin were retrieved from the NCBI database using Entrez. The follistatin protein of human was retrieved from the Swiss-Prot database. There are 344 amino acids and the molecular weight is 38,007 Da. The ProtParam analysis shows that the isoelectric point is 5.53 and the aliphatic index is 61.25. The hydropathicity is −0.490. The domains in FST protein are as follows: Pfam-B 5005 domain from 1 to 92; EGF-like subdomain from 93 to 116; Kazal 1 domain, occurred in three places, namely, 118–164, 192–239, and 270–316. There are 31 single-nucleotide polymorphisms (SNPs) for this gene. Some are nonsynonymous, some occur in the intron region, and some in an untranslated region. Two nonsynonymous SNPs, namely, rs11745088 and rs1127760, were taken for analysis. In the SNP rs11745088, the change is E152Q. Likewise, in rs1127760, the change is C239S. SIFT (Sorting Intolerant from Tolerant) showed positions of amino acids and the single letter code of amino acids that can be tolerated or deleterious for each position. There were six SNP results and each result had links to it. The dbSNP id, primary database id, and the type of mutation whether silent and if occurring in coding region are given as phenotype alterations. The FASTA format of protein was given to the nsSNP Analyzer tool, and the variation E152Q and C239S were given as inputs in the SNP data field. E152Q change was neutral and C239S causes disease. Using PANTHER for evolutionary analysis of coding SNPs, the protein sequence was given as input and analyzed for the E152Q and C239S SNPs for deleterious effect on protein function. The genetic association database results showed that FST gene SNPs are linked to PCOS coming under the disease class of metabolic disorders. The list of intronic and synonymous SNPs, with their nucleotide position, amino acid change information, and dbSNP link, is provided for further analysis.
Introduction
Polycystic ovarian syndrome (PCOS) is a common endocrine disorder that is found in ≈4% of women of reproductive ageCitation1 and results in reduced fertility and a sevenfold increased risk for type 2 diabetes mellitus.Citation2 The syndrome is characterized by hyperandrogenism and chronic anovulation. It is also associated with polycystic ovaries, hirsutism, obesity, and insulin resistance. The observation of familial aggregation of PCOSCitation3–Citation5 showed that it is consistent with a genetic basis for this disorder. The mode of inheritance is uncertain at this point, and the role of shared environmental factors such as diet and lifestyle in the presentation of the disease is unknown. Loci proposed and investigated as possible PCOS genes include CYP11A, the insulin gene, and a region near the insulin receptor gene.Citation6 The wide range of PCOS symptoms likely plays a significant role in the inability thus far to identify a specific gene mutation.
Single-nucleotide polymorphisms (SNPs) are DNA sequence variations that occur when a single nucleotide (A, T, C, or G) in the genome sequence is changed. Most SNPs, actually about two of every three SNPs, involve the replacement of cytosine (C) with thymine (T). SNPs occur every 100–300 bases along the human genome. SNPs are stable from an evolutionarily standpoint not changing much from generation to generation.
GGCTTCAGAATGGCC
GGCTTCAAAATGGCC
SNPs, which make up about 90% of all human genetic variation, occur every 100–300 bases along the 3-billion-base human genome. SNPs can occur in both coding (gene) and noncoding regions of the genome. Many SNPs have no effect on cell function, but scientists believe others could predispose people to disease or influence their response to a drug. SNP maps will help them to identify the multiple genes associated with such complex diseases as cancer, diabetes, vascular disease, and some forms of mental illness.
Several pathways have been implicated in the etiology of PCOS. These include the metabolic or regulatory pathways of steroid hormone synthesis,Citation7,Citation8 regulatory pathways of gonadotropin action,Citation9 the insulin-signaling pathway,Citation10–Citation12 and pathways regulating body weight.Citation13 Several genes from these pathways have been tested as candidate genes for PCOS.Citation7,Citation8,Citation14–Citation19 Although mutation analysis, linkage studies, and case-control association studies have been carried out with these candidate genes, evidence that any of them play a role in PCOS has not been replicated widely and is still inconclusive. These uncertainties are common in ‘complex’ genetic diseases, where identifying the contributing genes is made difficult by likely genetic heterogeneity, environmental contributions, and multiple etiologies. However, the mode of inheritance of PCOS has not been firmly established. Although some studies support a single dominant gene with high penetrance,Citation20–Citation22 others do not.Citation23 A study was conducted by Urbanek et alCitation24 on a carefully chosen collection of 37 candidate genes for linkage and association with PCOS or hyperandrogenemia in data from 150 families. The authors found that strongest evidence for linkage was with the follistatin gene.
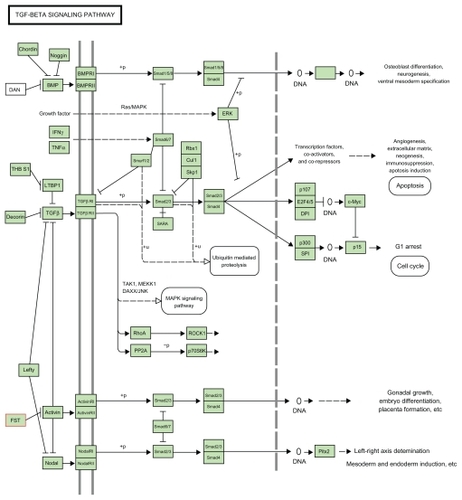
Strong evidence for a link between the follistatin gene and PCOS has been recently found in a well-designed large-scale study. The pathway in which follistatin gene is involved is shown in as in KEGG database: Follistatin binds to activin and affects its functions, for example, stimulation of follicle-stimulating hormone (FSH) synthesis and secretion. Thus, it may play a role in the functional impairment of the FSH–granulosa cell axis in PCOS. Several genes involved in the biosynthesis of androgen, and action of insulin and gonadotrophin have been examined as candidate genes for PCOS. These genes include those for the cholesterol side-chain cleavage enzyme (CYP11A), 17-hydroxylase/17,20-lyase (CYP17), insulin, insulin receptor, and LH. Two of them, CYP11A and the insulin gene, and variable number of tandem repeats, have been proposed as predisposing genetic factors contributing to PCOS. However, neither of them have been widely accepted as a major cause for this syndrome. Recently, in a well-designed large-scale study, Urbanek et alCitation24 tested for linkage and association between 37 candidate genes including those previously studied and PCOS. These genes were carefully selected from those involved in the action of androgen, gonadotrophin, and insulin, and also the regulation of obesity and energy. They found evidence for linkage only with the CYP11A and follistatin genes. However, only the linkage with follistatin gene remained significant after correction for multiple testing. Nevertheless, this study apparently confirmed previous findings which demonstrated the linkage of CYP11A gene with PCOS.
Follistatin is a single-chain glycosylated polypeptide that can bind to activin with high affinity and neutralize its biological action of stimulating the secretion of FSH and increasing FSHß mRNA levels and may therefore, arrest folliculogenesis. Indeed, overexpression of follistatin in the transgenic mice resulted in the suppression of both serum concentrations of FSH and ovarian folliculogenesis, similar to the clinical features commonly found in PCOS patients. Follistatin is expressed in numerous tissues including the ovary, pituitary, adrenal cortex, and pancreas. The human cDNAs encoding follistatin have been cloned. There is a single follistatin gene that can generate two mature mRNA transcripts by alternative splicing, thus encoding proteins of 315 (FS-315) and 288 (FS-288) amino acid residues, respectively. The FS-288, a carboxy-truncated variant with increased biological potency, was found to bind strongly to heparin sulphate proteoglycans of the cell membrane, whereas FS-315 had little or no such binding affinity. Furthermore, in the anterior pituitary cells, FS-288 was more potent in suppressing FSH release. This cell-associated protein was also found to accelerate the uptake of activin into pituitary cells, leading to an increase in its degradation by lysosomal enzymes and thus playing a role in the activin clearance system.
In view of this, it was thought that it would be worthwhile to explore the role of follistatin gene and its linkage with PCOS. The objectives of the work are retrieval of follistatin gene and protein, analysis of follistatin gene and protein, and SNP studies on follistatin to study SNP association with PCOS. Hence, in this work, SNPs in the FST gene were studied to determine if genetic variation is associated with susceptibility to PCOS or key phenotypic features of PCOS patients.
Materials and methods
The various methods, databases, tools, and software used for carrying out this project and their details are given below.
Sequence retrieval
Nucleotide – NCBI: The nucleotide sequence of human follistatin was retrieved from NCBI database using Entrez.
Protein – NCBI: The protein sequence of human follistatin was retrieved from NCBI database using Entrez.
Protein – Swiss-Prot: The follistatin protein of human was retrieved from Swiss-Prot database.
Sequence analysis
Protparam: The protein sequence of follistatin was given to Protparam search and analyzed.
Pfam: The protein sequence of follistatin was given to Pfam search and analyzed for the protein family.
SNP analysis
The follistatin gene and protein was subjected to various SNP analyses.
NCBI – dbSNP: The SNPs of human follistatin were retrieved from database. The list of NCBI SNP entries was obtained from genecard link.
SIFT (Sorting Intolerant from Tolerant): The follistatin protein sequences were submitted as input for the SIFT tool.
Single Amino Acid Polymorphism (SAAP): The SAAP is to link SNP to phenotype alterations. In this tool, the UniProt id P19883 was given in the search box.
nsSNP Analyzer: The FASTA format of protein was given to the nsSNP Analyzer tool and the variation E152Q and C239S were given as input in the SNP data field.
PANTHER – Evolutionary analysis of coding SNPs: The protein sequence was given as input and analyzed for the E152Q and C239S SNPs. Top of Form Estimates the likelihood of a particular nonsynonymous (amino acid changing) coding SNP to cause a functional impact on the protein. It calculates the substitution position-specific evolutionary conservation (subPSEC) score based on an alignment of evolutionarily related proteins, as described by Thomas et alCitation25 and Thomas and Kejariwal.Citation26 Multiple sequence alignment (MSA) position to view the column in the MSA where the substitution occurs was also carried out.
SNPs3D: SNPs3D is a Web site which assigns molecular functional effects of nonsynonymous SNPs based on structure and sequence analysis.
Results and discussion
Sequence retrieval
Nucleotide – NCBI: The nucleotide sequence of human follistatin was retrieved from NCBI database using Entrez. The accession number is BC004107 and there are totally 1326 base-pairs in the mRNA.
The sequence in FASTA format is given below: >gi|33871153|gb|BC004107.2| Homo sapiens follistatin, mRNA (cDNA clone MGC:10663 IMAGE:3688745), complete cds
Protein – NCBI: The protein sequence of human follistatin was retrieved from NCBI database using Entrez. The accession number is AAH04107 and there are totally 344 amino acids in the protein.
The protein in FASTA format is given below: >gi|13278648|gb|AAH04107.1| follistatin [Homo sapiens]
Protein – SWISSPROT: The follistatin protein of human was retrieved from SWISSPROT database and the accession number is P19883. There are 344 amino acids and the molecular weight is 38,007 Da. Out of the many genes follistatin was considered as a main candidate gene for PCOS. So, this gene was selected for study.
Follistatin was considered a candidate for the following reasons. It is an activin-binding protein that neutralizes the biological activity of activin in vivo and in vitro and is expressed in multiple tissues, including the ovary, pituitary, adrenal cortex, and pancreas. Activin, a member of the transforming growth factor-ß superfamily, modulates the production of androgens by ovarian thecal cells, the development of ovarian follicles, and the secretion of FSH by the pituitary and insulin by pancreatic ß-cells. Because follistatin inhibits the activity of activin, altered follistatin activity would be expected to affect follicular development, ovarian androgen production, pituitary FSH secretion, and insulin release. All these processes have been shown to be perturbed in PCOS.Citation27,Citation28
Sequence analysis
Protparam: The protein sequence of follistatin was given to Protparam search and analyzed.
Pfam: The protein sequence of follistatin was given to Pfam search and analyzed for the protein family.
FST_HUMAN (P19883): DE follistatin precursor (FS) (activin-binding protein). The protparam analysis showed that the isoelectric point is 5.53 and the aliphatic index is 61.25. The hydropathicity is −0.490. The domains in FST protein are Pfam-B 5005 domain from 1 to 92, EGF-like subdomain from 93 to 116, Kazal 1 domain occurring in three places, namely, 118–164, 192–239, and 270–316.
Family: Kazal_1 (PF00050): Alignment also includes a single domain from transporters in the OATP/PGT family (Swiss: P46721).
Interpro entry IPR002350: This family of Kazal inhibitors belongs to MEROPS inhibitor family I1, clan IA. They inhibit serine peptidases of the S1 family. These proteins contain between 1 and 7 Kazal-type inhibitor repeats. The structure of the Kazal repeat includes a large quantity of extended chain, two short α-helices and a three-stranded antiparallel β sheet. The inhibitor makes 11 contacts with its enzyme substrate: unusually, 8 of these important residues are hypervariable.
Altering the enzyme-contact residues, and especially that of the active site bond, affects the strength of inhibition and specificity of the inhibitor for particular serine proteases. The presence of this Pfam domain is usually indicative of serine protease inhibitors; however, Kazal-like domains are also seen in the extracellular part of agrins which are not known to be proteinase inhibitors.
Clan
This family is a member of clan Kazal (CL0005), which contains the following two members: Kazal_1 and Kazal_2.
Family: EGF-like_subdom (PF09120): Members of this family are reminiscent of EGF-like modules, as indicated by an identical disulphide connectivity. This subdomain comprises a stretch of residues in an extended conformation held in place by two disulphide linkages to two β strands. In follistatin, it is followed by a Kazal-like sequence: both are required for heparan sulphate binding.
Interpro entry IPR015204: This is an EGF-like domain, as indicated by an identical disulphide connectivity, which resembles the N-terminal domain of follistatin. It comprises a stretch of residues in an extended conformation held in place by two disulphide linkages to two β strands. In follistatin, it is followed by a Kazal-like sequence: both are required for heparan sulphate binding.
Clan
This family is a member of clan EGF (CL0001), which contains the following eight members: EGF, EGF-like_subdom, EGF_2, EGF_alliinase, EGF_CA, FOLN, Laminin_EGF, and Tme5_EGF_like.
SNP analysis
The follistatin gene and protein were subjected to various SNP analyses.
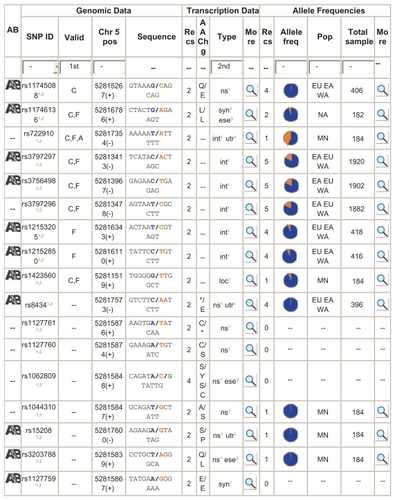
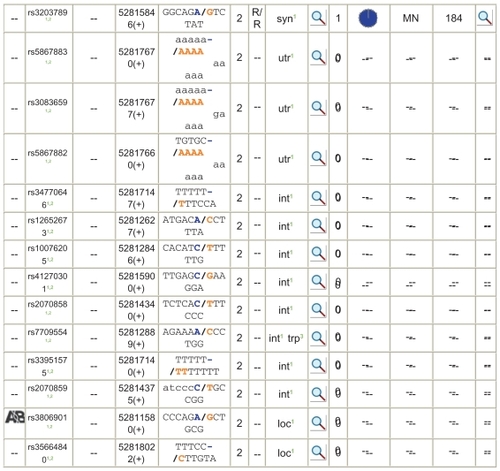
NCBI – dbSNP: The SNPs of human follistatin were retrieved from database. The list of NCBI SNP entries was obtained from genecard link. There are 31 SNPs for this gene and some are nonsynonymous, some occur in intron region, and some in untranslated region (). Two nonsynonymous SNPs, namely, rs11745088 and rs1127760, are taken for analysis. In the SNP rs11745088, the change is E152Q. That is E in position 152 is changed to Q. Likewise, in rs1127760 the change is C239S.
Recently, in a well-designed large-scale study, Urbanek et alCitation24 tested for linkage and association between 37 candidate genes including those previously studied and PCOS. These genes were carefully selected from those involved in the action of androgen, gonadotrophin, and insulin, and also the regulation of obesity and energy. They found evidence for linkage only with the CYP11A and follistatin genes. However, only the linkage with follistatin gene remained significant after correction for multiple testing.
Likewise, studies of Jones et alCitation28 in 173 Caucasian women suggest that polymorphism in the follistatin gene is associated with key androgenic phenotypes of PCOS. In their observation, the SNP rs3797297, located in intron 1 of the FST gene, was significantly associated with both FAI and SHBG. In both instances, post hoc analysis revealed that the main effect was due to subjects who were homozygous for rare A-allele of the polymorphism. The data for rs3797297 did not appear to be consistent with a codominant mode of expression at this locus. In subjects with two copies of the polymorphic allele at this locus, the FAI was well above the reference range, representing more severe hyperandrogenemia than in those women with a copy of the ancestral allele. Thus, a recessive effect appeared more likely. In the case of the SNP rs11745088, which was significantly associated with DHEA-S level, only two heterozygotes were identified for whom DHEA-S was available, with both these individuals having elevated DHEA-S. Although their study was larger than many molecular genetics studies of PCOS published to date, the power of the study remains relatively modest.
SIFT: The follistatin protein sequences were submitted as input for the SIFT tool. Amino acids that were not corresponding to its protein were indicated as ‘not tolerated’ means that the change might cause some deleterious effects. Forty-three sequences were selected to be closely related to our query sequence. At position 25R in the query sequence, 81% of the sequences have an amino acid appearing at this position. M, e, I, y, v, l, p, a, g, T, s, e, H, k, N, Q, D, R are predicted as tolerated and are observed in the alignment (capitalized). w and c are predicted to be deleterious because they have normalized probabilities <0.05 and none of these appear in the alignment (small letters). Amino acids are color coded: nonpolar, uncharged polar, basic, and acidic.
SAAP: SIFT showed positions of amino acids and the single letter code of amino acids that can be tolerated or deleterious for each position. The SAAP is to link SNP to phenotype alterations. In this tool, the UniProt id P19883 was given in the search box. The SAAP is to link SNP to phenotypic changes. There were six SNP results and each result had links to it. The dbSNP id, primary database id, and the type of mutation whether silent and if occurring in coding region are given phenotype alterations.
For 28 of the 37 candidate genes analyzed by Urbanek et al,Citation24 there is at least one polymorphic marker within 1 centiMorgan (cM) of the candidate gene, and for the remainder, the markers are 1–4 cM from the candidate gene. This proximity improves the power of the study because recombination is likely to be minimal.
nsSNP Analyzer: The FASTA format of protein was given to the nsSNP Analyzer tool and the variation E152Q and C239S was given as input in the SNP data field. E152Q change is neutral and the C239S causes disease.
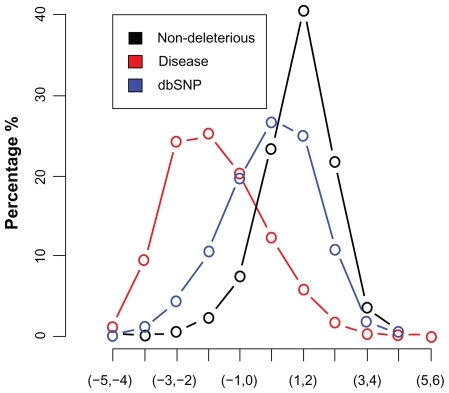
PANTHER – Evolutionary analysis of coding SNPs: The protein sequence was given as input and analyzed for the E152Q and C239S SNPs. The probability that a given variant will cause a deleterious effect on protein function is estimated by Pdeleterious such that a subPSEC score of −3 corresponds to a Pdeleterious of 0.5.
SNPS3D: The disease (HGMD) and nondeleterious dataset were used to train and test the model. ‘0’ was used as the threshold of classification. A SNP with a negative support vector machine (SVM) score is classified as damaging SNP. A SNP with a positive SVM score is classified as neutral SNP. A higher score (±) relates a higher confidence in prediction. A score >0.5 is considered to be significant (). The results showed that FST gene SNPs are linked to PCOS coming under the disease class of metabolic disorders.
Activin, a member of the transforming growth factor-b superfamily, and follistatin are expressed in several tissues, such as ovary, pituitary, adrenal cortex, and pancreas. This similar pattern of tissue distribution suggests that follistatin is likely to be an auto-/paracrine regulator of the hormonal effects of activin within the tissues. In particular, activin promotes ovarian follicular development, enhances LH binding sites and progesterone production, playing a role in preventing premature luteinization of the ovarian follicles, inhibits thecal cell androgen production, and increases pituitary FSH secretion and insulin secretion by pancreatic b-cells. An increase in circulating level or biological activity of follistatin may, therefore, cause follicular development to be arrested, the process of follicular luteinization or atresia to be favored, ovarian androgen production to be increased, FSH circulating levels to be reduced, and insulin release to be impaired. These effects are all characteristic features of PCOS.Citation29
Follistatin is a single-chain glycoprotein that primarily acts to regulate the activity of activin, which is responsible for ovarian follicular development, inhibition of theca cell androgen production and increases in both pituitary FSH secretion and pancreatic insulin secretion.Citation30 Primarily synthesized in the granulosa cells of the ovarian antral follicle, follistatin mRNA increases within the dominant follicle during development and declines during the atretic process.Citation31 Follistatin acts to suppress aromatase activity in the granulosa cell and also LH-stimulated progesterone release from thecal cells.Citation32 The overexpression of follistatin in mice has been shown to result in arrested ovarian follicular development and reduced levels of FSH, both key phenotypes of PCOS.Citation27
The list of intronic and synonymous SNPs, with their nucleotide position, amino acid change information, and dbSNP link, is provided for further analysis. Researchers were unable to detect any mutation of the activating or inhibiting type in the entire coding region of follistatin gene in 64 patients with PCOS. Therefore, mutations in the coding regions of the follistatin gene may not be a common cause of PCOS in the population studied. However, it is possible that mutations may reside in the regulatory region of the gene, which should be screened once its sequence is known. Furthermore, it would be of great interest to investigate the presence of mutations in PCOS patients in other ethnic populations, especially of European origin, as it was in this population that the linkage between the follistatin gene and PCOS was established.Citation33
Strong evidence for a link between the follistatin gene and PCOS has been recently found in a well-designed large-scale study. Odunsi and KiddCitation34 are of the opinion that overexpression of follistatin will be expected to lead to increased ovarian androgen production and reduction in circulating FSH levels, which are features of PCOS. The genetic association database results show that FST gene SNPs are linked to PCOS coming under the disease class of metabolic disorders. The list of intronic and synonymous SNPs, with their nucleotide position, amino acid change information, and dbSNP link, is provided for further analysis.
The authors thus conclude that there is an association between FST gene SNPs and polycystic ovarian syndrome.
Disclosure
The authors report no conflict of interest in this work.
References
- KnochenhauerESKeyTJKahsar-MillerMWaggonerWBootsLRAzzizRPrevalence of the polycystic ovary syndrome in unselected black and white women of the southeastern United States: a prospective studyJ Clin Endocrinol Metab199883307830829745406
- LegroRSKunselmanARDodsonWCDunaifAPrevalence and predictors of risk for type 2 diabetes and impaired glucose tolerance in polycystic ovary syndrome: a prospective, controlled study in 254 affected womenJ Clin Endocrinol Metab1999841651699920077
- CooperHESpellacyWNPremKACohenWDHereditary factors in the Stein-Leventhal syndromeAm J Obstet Gynecol196810037138715782458
- GivensJRFamilial polycystic ovarian diseaseEndocrinol Metab Clin North Am1988177717833058473
- LegroRSSpielmanRUrbanekMDriscollDStraussJFDunaifAPhenotype and genotype in polycystic ovary syndromeRecent Prog Horm Res1998532172569769710
- LegroRSStraussJFMolecular progress in infertility: polycystic ovary syndromeFertil Steril200278569576
- CareyAHWaterworthDPatelKPolycystic ovaries and premature male pattern baldness are associated with one allele of the steroid metabolism gene CYP17Hum Mol Genet19943187318767849715
- GharaniNWaterworthDMBattySAssociation of the steroid synthesis gene CYP11a with polycystic ovary syndrome and hyperandrogonismHum Mol Genet199763974029147642
- FranksSPolycystic ovary syndromeN Engl J Med19953338538617651477
- CiaraldiTPel-RoeiyAMadarZReichertDOlefskyJMYenSSCellular mechanisms of insulin resistance in polycystic ovarian syndromeJ Clin Endocrinol Metab1992755775831322430
- DunaifASegalKRShelleyDRGreenGDobrjanskyALicholaiTEvidence for distinctive and intrinsic defects in insulin action in polycystic ovary syndromeDiabetes199241125712661397698
- DunaifAXiaJBookC-BSchenkerETangZExcessive insulin receptor serine phosphorylation in cultured fibroblasts and in skeletal muscle: a potential mechanism for insulin resistance in the polycystic ovary syndromeJ Clin Invest1995968018107635975
- KiddyDSHamilton-FairleyDBushAImprovement in endocrine and ovarian function during dietary treatment of obese women with polycystic ovary syndromeClin Endocrinol199236105111
- O’RahillySChoiWHPatelPTurnerRCFlierJSMollerDEDetection of mutations in the insulin receptor gene in non-insulin dependent diabetic patients by analysis of single-stranded conformation polymorphismsDiabetes1991407777822040394
- TaylorSICamaAAcciliDMutations in the insulin receptor geneEndocr Rev1992135665951330507
- ConwayGSAveyCRumsbyGGenetics: the tyrosine kinase domain of the insulin receptor gene is normal in women with hyperinsulinaemia and polycystic ovary syndromeHum Reprod19949Suppl 9168116837836517
- KrookAKumarSLaingIBoultonAJWassJAO’RahillySMolecular scanning of the insulin receptor gene in syndromes of insulin resistanceDiabetes1994433573688314008
- SorbaraLRTangZCamaAAbsence of insulin receptor gene mutations in three women with the polycystic ovary syndromeMetabolism199443156815747990713
- KrookAO’RahillySMutant insulin receptors in syndromes of insulin resistanceBaillieres Clin Endocrinol Metab199610971228734453
- CareyAHChanKLShortFWhiteDWilliamsonRFranksSEvidence for a single gene effect causing polycystic ovaries and male pattern baldnessClin Endocrinol199338653658
- LegroRSDriscollDStraussJFFoxJDunaifAEvidence for a genetic basis for hyperandrogenemia in the polycystic ovary syndromeProc Natl Acad Sci U S A19989514956149609843997
- GovindAObhraiMClaytonRPolycystic ovaries are inherited as an autosomal dominant trait: analysis of 29 polycystic ovary syndrome and 10 control familiesJ Clin Endocrinol Metab199984Suppl 138439920059
- JahanfarSEdenJAWarrenPSeppalaMNguyenTVA twin study of polycystic ovary syndromeFertil Steril1995634784867531655
- UrbanekMLegroRSDriscollDAThirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatinProc Natl Acad Sci U S A1999968573857810411917
- ThomasPDCampbellMCKejariwalAPANTHER: a library of protein families and subfamilies indexed by functionGenome Res2003132129214112952881
- ThomasPDKejariwalACoding single-nucleotide polymorphisms associated with complex vs Mendelian disease: evolutionary evidence for differences in molecular effectsProc Natl Acad Sci U S A200410143153981540315492219
- UrbanekMWuXVickeryKRAllelic variants of the follistatin gene in polycystic ovary syndromeJ Clin Endocrinol Metab200085Suppl 124455446111134093
- JonesMRWilsonSGMullinBHMeadRWattsGFStuckeyBGAPolymorphism of follistatin gene in polycystic ovary syndromeMol Hum Reprod200713423724117284512
- AsteriaCIdentification of follistatin as a possible trait-causing gene in polycystic ovary syndromeEur J Endocrinol.2000143Suppl 446746911022191
- CalvoRVilluendasGSanchoJSan MillanJLEscobar-MorrealeHFRole of the follistatin gene in women with polycystic ovary syndromeFertil Steril2001751020102311334920
- LinS-YMorrisonJRPhillipsDJde KresterDMRegulation of ovarian function by the tgf-ß superfamily and follistatinReproduction200312613314812887271
- PhillipsDde KresterDMFollistatin: a multifunctional regulatory proteinFront Neuroendocrinol1998192873229799587
- LiaoWXRoyACNgSCPreliminary investigation of follistatin gene mutations in women with polycystic ovary syndromeMol Hum Reprod20006Suppl 758759010871644
- OdunsiKKiddKKA paradigm for finding genes for a complex human trait: polycystic ovary syndrome and follistatinProc Natl Acad Sci U S A.199996Suppl 158315831710411866