Abstract
Background
The human progesterone receptor (hPR) belongs to the steroid receptor family. It may be found as monomers (A and B) and or as a dimer (AB). hPR is regarded as the prognostic biomarker for breast cancer. In a cellular dimer system, AB is the dominant species in most cases. However, when a cell coexpresses all three isoforms of hPR, the complexity of the action of this receptor increases. For example, hPR A suppresses the activity of hPR B, and the ratio of hPR A to hPR B may determine the physiology of a breast tumor. Also, persistent exposure of hPRs to nonendogenous ligands is a common risk factor for breast cancer. Hence we aimed to study progesterone and some nonendogenous ligand interactions with hPRs and their molecular docking.
Methods and results
A pool of steroid derivatives, namely, progesterone, cholesterol, testosterone, testolectone, estradiol, estrone, norethindrone, exemestane, and norgestrel, was used for this in silico study. Dockings were performed on AutoDock 4.2. We found that estrogens, including estradiol and estrone, had a higher affinity for hPR A and B monomers in comparison with the dimer, hPR AB, and that of the endogenous progesterone ligand. hPR A had a higher affinity to all the docked ligands than hPR B.
Conclusion
This study suggests that the exposure of estrogens to hPR A as well as hPR B, and more particularly to hPR A alone, is a risk factor for breast cancer.
Introduction
The human progesterone receptor (hPR), like other members of the steroid receptor family of proteins, is a ligand-induced transcription factor which mediates the effects of progesterone.Citation1–Citation3 Progesterone is well known for its critical role in regulation of the normal physiology of the ovary, uterus, mammary gland, as well as brain development during childhood. Progesterone also plays an important role in the maintenance of the cardiovascular, central nervous, and skeletal systems.Citation4–Citation7 The importance of the hPR can therefore be understood from its extensive involvement in human physiological processes.
hPR is comprised of specific functional domains, including the central DNA-binding domain and a carboxyl-terminal ligand-binding domain. The central DNA-binding and ligand-binding domains are the sites of hPR activity. In addition to these domains, hPR have many elements with activator and inhibitory functions enhancing and repressing the transcriptional activation of hPR by their interaction with different transcriptional coregulators.3,8–12 Cytoplasmic hPR have been found as multiprotein chaperone complexes which facilitate their inactive conformation, whereby a ligand can bind with hPR.Citation13,Citation14
Two distinct isoforms (hPR A and hPR B) of hPR have been reported previously, and differ by an additional amino acid at position 164 in hPR B. These differences arise as a result of either alternate initiation of translation by the same mRNA or by transcription from alternate promoters within the same gene.Citation15
Our understanding of the mechanisms underlying the different activities of these two isoforms of human hPR is limited. However, structural and functional studies indicate that the hPR B isoform contains an additional domain, AF3, that accounts for the transcriptional activity of hPR B by suppressing the activity of an inhibitory domain contained within the sequences common to hPR A and hPR B.Citation11,Citation16,Citation17 The evidence suggests that the two isoforms acquire different conformations within the cell, and hence interact with distinct coregulators, ie, the coregulators of hPR A are different from those of hPR B.Citation18–Citation20
Much of our knowledge regarding the structure and function of hPR A and hPR B comes from models of single hPR isoform expression and models where the hPR homodimer is the dominant molecular species. However, cell types which coexpress the hPR isoforms may possibly have all three kinds of molecules, ie, hPR A, hPR B, and hPR AB, which increases the complexity of the action of hPR.
hPR is a ligand-activated receptor, and its activation/deactivation is associated with the pathogenesis of many diseases, including breast cancer.Citation21,Citation22 Hence, the specificity of hPR with regard to interaction with its ligand (progesterone) is an important area of research. It has been reported that prolonged exposure to certain nonendogenous ligands, particularly estrogens (which are steroid derivatives) is the strongest risk factor for breast cancer.Citation23,Citation24 In the absence of progesterone, the hPR A:hPR B ratio influences the biology of estrogen receptor-positive tumors and their response to treatment, and hPR A isoforms are functionally dominant in progesterone-deficient states. This explains why PR A-rich tumors are particularly aggressive.Citation25 We hypothesized that hPR A, hPR B, and hPR AB have different affinities for different ligands, based on conditions such as the availability of progesterone and the concentration of hPR monomers and dimers, which may be a possible cause for the development of breast cancer. The aim of our study was to test this hypothesis and find the affinities of the different steroid derivatives for hPRs. Hence, other steroids that can trigger the development of breast cancer by forming a ligand receptor complex with hPR can be predicted. A battery of steroid derivatives, namely, progesterone (DB00396), cholesterol (DB04540), testosterone (DB00624), testolectone (DB00894), estradiol (DB00783), estrone (DB00655), norethindrone (DB00717), exemestane (DB00990), and norgestrel (DB00506), was used for this in silico study ().
Methods and materials
Software, data sources, and experiments
All the software used in this study is freely available for academic use. provides the source of the data, and lists the software and online servers used. The Protein Data Bank (www.rcsb.org) is a World Wide Web repository where data are processed and distributed in the form of three-dimensional biological macromolecular structures.Citation26 The Protein Data Bank now has more than 20,000 structures determined by x-ray diffraction. Protein structures can be accessed and downloaded using keywords or the Protein Data Bank alphanumeric file name.
Table 1 Sources of data used in this study
Table 2 Software and online servers and corresponding URLs
The Drug Bank (http://redpoll.pharmacy.ualberta.ca/drugbank/) database is a unique bioinformatics and cheminformatics resource that combines detailed drug (ie, chemical, pharmacological, pharmaceutical) data with comprehensive drug target information (ie, sequence, structure, pathway).Citation27 The database contains nearly 4300 drug entries, including more than 1000 small molecule drugs approved by the US Food and Drug Administration (FDA), 113 FDA-approved biotechnologic (protein/peptide) drugs, 62 nutraceuticals, and more than 3000 experimental drugs. Additionally, more than 6000 protein (ie, drug target) sequences are linked to these drug entries. Each drug card entry contains more than 80 data fields, with half of the information concerning drug and chemical data and the other half concerning drug target or protein data.
PyMOL is a Python-based visualization program (version1.1r1; Delano Scientific 2006, www.pymol.org).Citation28 PyMOL was used to obtain graphical representations of proteins in the Protein Data Bank and the amino acid residues are in contact with the ligand. PyMOL tools can measure distances as well as identify different amino acid residues of receptors that interact with the ligands. Docking results were also analyzed by PyMOL.
AutoDock 4.2 docking software (Scripps Research Institute, www.scripps.edu) and AutoDockTools was used to perform the docking experiments.Citation29–Citation31 AutoDockTools is an accessory program that allows the user to interact with AutoDock from a graphic user interface. AutoDock is a suite of automated docking tools designed to predict how small molecules/ligands bind to a receptor/protein of known three-dimensional structure. AutoDock consists of three separate programs, ie, AutoDock, which docks the ligand to a set of grids describing the target protein, AutoGrid, which precalculates these grids, and AutoTors, which sets up bonds which will be treated as rotatable within the ligand. We calculated the binding energy of the different ligands for hPR A, hPR B, and hPR AB, and the molecular docking process was performed according to the method described by Mukharjee and Majumder.Citation32 The details of the procedure are as follows.
Preparing hPRs and ligand files for AutoDock
The Protein Data Bank files downloaded from the World Wide Web repository are often not perfect for docking studies because of missing hydrogen atoms, presence of multiple molecules, added waters, and related problems. Using the AutoDockTools graphic user interface, the files were prepared as follows.
For the hPRs file, all heteroatoms in the Protein Data Bank files were removed. Three separate Protein Data Bank files were then prepared for hPR A, hPR B, and hPR AB, and saved with an extension of .pdb. All three Protein Data Bank files were first read in AutoDockTools, any added waters removed, and polar hydrogens added. AutoDockTools was then used to check if the molecule had charges and, if not, AutoDockTools determined whether the molecule was a peptide (ie, whether all of the names of its residues appeared in the standard set of 20 commonly occurring amino acids). If the molecule was found to be a peptide, Kollman charges were added, otherwise Gasteiger charges were added. Finally, solvation parameters were added, and the files were saved with .pdbqs extension (where q and s represent charge and solvation, respectively).
Ligand files
Protein Data Bank files of ligands obtained from the Drug Bank, were read in AutoDockTools, modified as necessary, and saved using .pdbqs extension. AutoDockTools was then used for automatic calculation of the best root, ie, the fixed portion of the ligand from which rotatable branches sprout. We also determined the rotatable bonds in the ligand, making all amide bonds nonrotatable, and set the number of active torsions to the least number of atoms. The ligand files were then saved with ligand.out.pdbq extension (q representing charge).
Grid parameter files
For the calculation of interaction energy, parameters were set to create a grid (ie, a three-dimensional box) capable of enclosing hPRs molecules. The grid volume was large enough to allow the ligand to rotate freely, even with its most fully extended conformation. Grid parameters were stored in a grid parameter file using molecule.gpf extension.
AutoGrid4 program
The AutoGrid4 program was run, creating a map for every type of atom in the ligand. For example, a molecule having carbon, nitrogen, oxygen, and hydrogen maps was created as a molecule.C.map, molecule.N.map, molecule.O.map, and molecule.H.map. These grid maps were saved in ASCII format for readability by AutoDock. AutoGrid also generates a corresponding output of the macromolecular file with the extension molecule.glg.
Docking parameter file
The docking parameter file and other properties defined for the ligand was created. A docking parameter file instructs AutoDock on which ligand to move and the map files to use. AutoDock’s search methods include the Monte Carlo simulated annealing method, the genetic algorithm, local search, and the hybrid genetic algorithm with local search (GALS). The GALS is also considered as the Lamarckian genetic algorithm because the offsprings are allowed to inherit the local search adaptations of their parents. GALS was the chosen algorithm for hPR and ligand interaction analysis.
AutoDock program
Finally, the AutoDock program was run from the AutoDock graphic user interface, and the docked ligand files (.dlg extension) were used for this study. The .dlg files were read in Autodock as well as in PyMOL to calculate the binding energies in the docked ligand-protein complexes.
Results
Rigorous docking experiments were done to assess the cross-reactivity of various steroid derivatives (ligands) with hPR monomers and dimers. The ligands, receptors, and their corresponding docking results are listed in , and receptor-ligand interactions demonstrated in , , and . In these docking experiments, all selected ligands were able to bind with hPR A. Estradiol showed the highest binding affinity (ΔG kcal/mol) at −3.68e+06, followed by estrone, progesterone, and cholesterol (3.38e+06, −3.54e+06, and −2.19e+06 kcal/mol, respectively). Testosterone, testolactone, norethindrone, exemestane, and norgestrel exhibited binding affinities in the range of −8.73 to −7.14 kcal/mol. Ligand interactions with hPR B showed lower binding affinity than with hPR A. A similar pattern of binding affinity was found for ligands with hPR B, except for progesterone and cholesterol. Estradiol had the highest binding affinity at −3.64e+6 kcal/mol followed by estrone, cholesterol, and progesterone (3.19e+6, −2.53e+6, and 2.05e+6 kcal/mol, respectively). Testosterone, testolactone, norethindrone, exemestane, and norgestrel exhibited binding affinities in the range of −5.86 to −4.75 kcal/mol. All the ligand interactions with hPR homodimer (AB) had similar binding affinity, except for cholesterol, which had a binding affinity of −1.86e+6 kcal/mol, which was the highest binding affinity among all the ligand interactions with hPR AB. Progesterone had a binding affinity of −10.82 kcal/mol, and the remainder of the ligand binding affinities were in the range of −9.82 to −8.50 kcal/mol.
Figure 2 Progesterone (A), cholesterol (B), testosterone (C), testolactone (D), estradiol (E), estrone (F), norethindrone (G), exemestane (H), and norgestrel (I) docked onto the human progesterone receptor A (hPR A, green) in its lowest energy-docked conformation. Ligand molecules are shown in red.
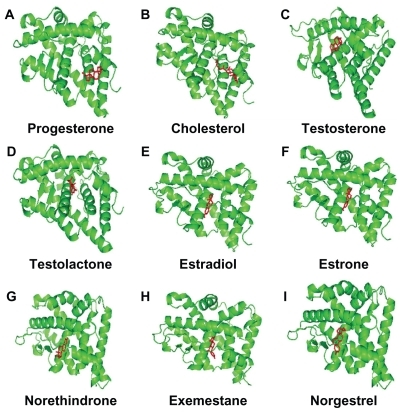
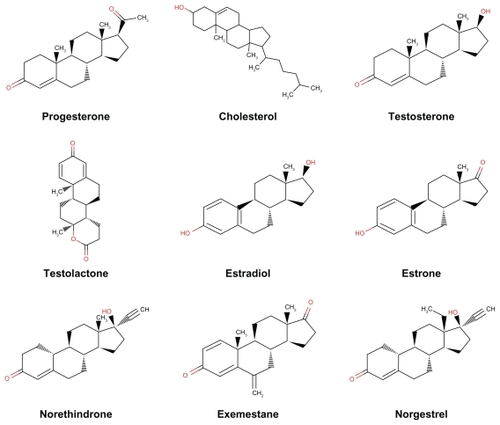
Figure 3 Progesterone (A), cholesterol (B), testosterone (C), testolactone (D), estradiol (E), estrone (F), norethindrone (G), exemestane (H), and norgestrel (I) docked onto the human progesterone receptor A (hPR B green) in its lowest energy-docked conformation. Ligand molecules are shown in red.
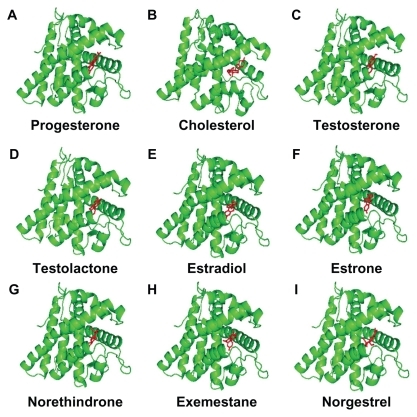
Figure 4 Progesterone (A), cholesterol (B), testosterone (C), testolactone (D), estradiol (E), estrone (F), norethindrone (G), exemestane (H), and norgestrel (I) docked onto the human progesterone receptor A (hPR AB green) in its lowest energy-docked conformation. Ligand molecules are shown in red.
Table 3 Docking results of steroid derivative (ligand) molecules at human progesterone receptor chains (A, B, and AB, blind docking)
Using this molecular modeling study, we found that the estrogens, estradiol and estrone, had higher affinity for the hPR monomers A and B. Therefore, we undertook binding site analyses for hPR A, B, and AB, for estradiol and estrone. summarizes the amino acid residues involved in the binding of estradiol and estrone with hPR A, hPR B, and AB. Only the HIS 93 residue of hPR A created a single hydrogen bond (H-bond) with estradiol and estrone (bond length 2.0 and 2.3 Å, respectively). In the case of hPR B, estradiol created two H-bonds, one with TRP 78 (2.2 Å) and the other with ARG 89 (2.0 Å), whereas estrone made three H-bonds, two with GLN 48 (2.2 and 2.2 Å) and one with ARG 89 (2.1 Å). Interestingly, hPR AB was not involved in the formation of any H-bond with estrone and only ASP 205 of hPR AB made a single H-bond with estradiol (1.7 Å). The rest of the amino acid residues listed in were involved in hydrophobic interactions through their side chains.
Table 4 Amino acid residues involved in binding of estradiol and estrone with human progesterone receptors A, B, and AB
Discussion
Specificity in biomolecular interactions is determined mainly by noncovalent forces like Coulombic electrostatic forces, hydrogen bonds, London’s dispersive forces, and van der Waals forces. In addition, the structure of biomolecules is a function of forces among the constituent structural elements, as well as environmental forces. Binding affinity is inversely proportional to free Gibbs energy. To find the specificity of hPR for different steroid derivatives, monomers and dimers of hPR were studied separately because it had been found in vivo that there is a difference in the binding pattern of ligands to hPR A and B. They adopt distinct conformations in cells, as well as having different signaling responses.Citation18–Citation20,Citation33–Citation35 Earlier studies of elephant and hamster uterine progesterone receptors reported that there are many natural and synthetic steroid derivatives which are nonendogenous ligands and have higher relative binding affinity with progesterone receptors as compared with progesterone.Citation36,Citation37 Similarly, progesterone is the endogenous ligand for hPR, but in this in silico study, where there was one-to-one correspondence between hPRs and ligands, as well as their flexible docking, we found that estradiol has more affinity with hPR A than progesterone, and estrone had less affinity with hPR A as compared with progesterone, whereas cholesterol has a higher Gibbs energy value compared with progesterone. In the case of hPR B, estrone, estradiol, and cholesterol molecules had higher affinity with hPR B than did progesterone. In general, the binding energy for the ligands with hPR A was considerably higher than that of hPR B. This supports the view that, in the presence of hPR A and hPR B monomers in a cellular system, hPR A can exert an inhibitory effect on the transactivator behavior of hPR B.Citation16,Citation17 Our finding is also in agreement with earlier studies showing that hPR A-rich tumors are more aggressive.Citation25 The higher binding affinity of ligands like estradiol and estrone for hPR A than for hPR B is supported by the previous study which showed that cumulative exposure of estradiols to hPR is the most common risk factor for the development of breast cancer.
Prolonged lifetime exposure to estrogens, especially estrone and estradiol, has long been linked to the promotion and progression of breast cancer because of their physiological action on the mammary gland.Citation23,Citation24 Another in vitro study also reports significantly increased stimulation of the progesterone receptor by estrone,Citation38 although there is a lot of evidence suggesting that binding of ligands with hPRs and their transcriptional activity is also a function of phosphorylation of C-terminal amino acid residues as well as corresponding conformational changes in hPRs.Citation39–Citation41 Due to the limitations of this in silico molecular modeling study, we did not consider different conformational changes of hPRs due to their differential phosphorylation inside the cells, and further crystallographic studies are required to confirm our findings. Our results may contribute an insight into the risk of breast cancer due to nonendogenous ligands, especially estrogens that bind with hPRs, although the higher binding energy of cholesterol with hPR AB cannot be explained by this study. However, it can be predicted that, in the presence of a deficiency of progesterone or an abundance of cholesterol, formation of a cholesterol-hPR AB complex may be favored at equilibrium, and may be another risk factor for breast cancer. This observation is in agreement with earlier studies reporting that increased total cholesterol levels are associated with development of breast cancer which is resistant to certain drugs, eg, tamoxifen.Citation42–Citation44
Conclusion
The absence of higher binding affinity of hPRs with progesterone in comparison with estradiol and estrone in a cellular or animal model can be explained by ligand receptor interaction kinetics and also by study of the physiological fate of such ligands in animal and cellular systems. This study may enhance our understanding of the specificity in binding of steroid derivatives to hPR. Our findings advocate that estradiol and estrone binds with hPR A and hPR B monomers with higher affinity in comparison to progesterone. Such interactions cause aggressive tumor development. Hence, our study may be helpful in understanding why the hPR A:hPR B ratio and prolonged exposure of nonendogenous ligands, particularly estrogens, to hPRs in the presence and absence of progesterone, is a risk factor for breast cancer.
Acknowledgments
The authors are grateful to Dr MSM Jaabir for advice during this study. We also extend our thanks to the administrative staff of the Department of Bioinformatics, Jamal Mohammed College, Bharathidasn University, India, and the Department of Food Science and Nutrition, King Saud University, Saudi Arabia, for allowing the laboratory facility to carry out this work.
Disclosure
The authors report no conflicts of interest in this work.
References
- EscrivaHBertrandSLaudetVThe evolution of the nuclear receptor superfamilyEssays Biochem200440112615242336
- EvansRMThe steroid and thyroid hormone receptor superfamilyScience199824048548898953283939
- McEwanIJNuclear receptors: One big familyMethods Mol Biol2009505131819117136
- GrahamJDClarkeCLPhysiological action of progesterone in target tissuesEndocr Rev19971845025199267762
- GrahamJDClarkeCLExpression and transcriptional activity of progesterone receptor A and progesterone receptor B in mammalian cellsBreast Cancer Res20024518719012223122
- LiXLonardDMO’MalleyBWA contemporary understanding of progesterone receptor functionMech Ageing Dev200412510–1166967815541762
- MotePAGrahamJDClarkeCLProgesterone receptor isoforms in normal and malignant breastErnst Schering Found Symp Proc200717710718540569
- EdwardsDPThe role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptorsJ Mammary Gland Biol Neoplasia20005330732414973393
- McKennaNJLanzRBO’MalleyBWNuclear receptor coregulators: Cellular and molecular biologyEndocr Rev199920332134410368774
- McKennaNJO’MalleyBWCombinatorial control of gene expression by nuclear receptors and coregulatorsCell2002108446547411909518
- SartoriusCAMelvilleMYHovlandARTungLTakimotoGSHorwitzKBA third transactivation function (AF3) of human progesterone receptors located in the unique N-terminal segment of the B-isoformMol Endocrinol1994810134713607854352
- VegetoEShahbazMMWenDXGoldmanMEO’MalleyBWMcDonnellDPHuman progesterone receptor A form is a cell- and promoter-specific repressor of human progesterone receptor B functionMol Endocrinol1993710124412558264658
- SmithDFFaberLEToftDOPurification of unactivated progesterone receptor and identification of novel receptor-associated proteinsJ Biol Chem19902657399640032303491
- PrattWBGalignianaMDMorishimaYMurphyPJRole of molecular chaperones in steroid receptor actionEssays Biochem200440415815242338
- KastnerPKrustATurcotteBTwo distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and BEMBO J199095160316142328727
- Abdel-HafizHTakimotoGSTungLHorwitzKBThe inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepressionJ Biol Chem200227737339503395612114521
- TungLAbdel-HafizHShenTProgesterone receptors (PR)-B and -A regulate transcription by different mechanisms: AF-3 exerts regulatory control over coactivator binding to PR-BMol Endocrinol200620112656267016762974
- BainDLFrandenMAMcManamanJLTakimotoGSHorwitzKBThe N-terminal region of the human progesterone A-receptor: Structural analysis and the influence of the DNA binding domainJ Biol Chem2000275107313732010702302
- BainDLFrandenMAMcManamanJLTakimotoGSHorwitzKBThe N-terminal region of human progesterone B-receptors: Biophysical and biochemical comparison to A-receptorsJ Biol Chem200127626238252383111328821
- TetelMJGiangrandePHLeonhardtSAMcDonnellDPEdwardsDPHormone-dependent interaction between the amino-and carboxyl- terminal domains of progesterone receptor in vitro and in vivoMol Endocrinol199913691092410379890
- FuquaSAWCuiYLeeAVOsborneCKHorwitzKBInsights into the role of progesterone receptors in breast cancerJ Clin Oncol200523493193215681552
- RevillionFLindetCLhotellierVHornezLBonneterreJPeyratJRelationships between progesterone receptor isoforms and the HER receptors and ligands network in 299 primary breast cancersCancer Res20096924 Suppl Abstr 4150
- ClemonsMGossPEstrogen and the risk of breast cancerN Engl J Med2001344427628511172156
- DorganJFBaerDJAlbertPSHormones and the alcohol-breast cancer association in postmenopausal womenJ Natl Cancer Inst200193971071511333294
- JacobsenMBSchittoneSARicherJKHorwitzKBProgesterone-independent effects of human progesterone receptors (PRs) in estrogen receptor-positive breast cancer: PR isoform-specific gene regulation and tumour biologyMol Endocrinol200519357458715563544
- BermanHMWestbrookJFengZThe Protein Data BankNucleic Acids Res200028123524210592235
- WishartDSKnoxCGuoCADrug Bank: A comprehensive resource for in silico drug discovery and explorationNucleic Acids Res2006134D66867216381955
- DeLanoWLThe PyMol Molecular Graphics System2002 Available at: http://www.pymol.orgAccessed February 7, 2011
- MorrisGMGoodsellDSHallidayRSAutomated docking using a Lamarckian genetic algorithm and an empirical binding free energy functionJ Comput Chem1998191416391662
- MorrisGMGoodsellDSHueyRMolecular Graphics Laboratory, 2001 Available at: http://autodock.scripps.edu/faqs-help/manual/autodock-3-user-s-guideAccessed February 7, 2011
- HueyRMorrisGMUsing AutoDock4 with AutoDockTools: A tutorialLa Jolla, CAThe Scripps Research Institute, Molecular Graphics Laboratory2008
- MukherjeeSMajumderDComputational molecular docking assessment of hormone receptor adjuvant drugs: Breast cancer as an examplePathophysiology2009161192919147336
- GiangrandePHKimbrelEAEdwardsDPMcDonnellDPThe opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor bindingMol Cell Biol20002093102310510757795
- HeneghanAFConnaghan-JonesKDMiuraMTBainDLCoactivator assembly at the promoter: Efficient recruitment of SRC2 is coupled to cooperative DNA binding by the progesterone receptorBiochemistry20074639110231103217845055
- Molenda-FigueiraHAMurphySDSheaKLSteroid receptor coactivator-1 from brain physically interacts differentially with steroid receptor subtypesEndocrinology2008149105272527918566116
- GreylingMDvan AardeRJPotgieterHCLigand specificity of uterine oestrogen and progesterone receptors in the subadult African elephant Loxodonta africanaJ Reprod Fertil199710921992049155728
- WilksJWSpilmanCHCampbellJASteroid binding specificity of the hamster uterine progesterone receptorSteroids19803566977067404605
- LanzoneANguyenBLPasqualiniJRUptake, receptor and biological response of estrone in the fetal uterus of guinea pigHorm Res19831731681806852778
- WeigelNLBeckCAEstesPALigands induce conformational changes in the carboxyl-terminus of progesterone receptors which are detected by a site-directed antipeptide monoclonal antibodyMol Endocrinol1992610158515971448113
- NarayananREdwardsDPWeigelNLHuman progesterone receptor displays cell cycle-dependent changes in transcriptional activityMol Cell Biol20052582885289815798179
- ClemmDLShermanLBoonyaratanakornkitVSchraderWTWeigelNLEdwardsDPDifferential hormone-dependent phosphorylation of progesterone receptor A and B forms revealed by a phosphoserine site-specific monoclonal antibodyMol Endocrinol2000141526510628747
- Hasija BaggaHAlterations of serum cholesterol and serum lipoprotein in breast cancer of womenInt J Clin Biochem20052016166
- RigginsRBLanJPKlimachUERRγ mediates tamoxifen resistance in novel models of invasive lobular breast cancerCancer Res200868218908891718974135
- RigginsRBLanJPDunderdaleJZwartAClarkeRERRγ/AP1 signaling drives cholesterol production and tamoxifen resistance in breast cancerAmerican Association for Cancer Research Annual MeetingDenver, COApril 18–22, 2009