Abstract
The lack of complete concordance of autoimmune disease in identical twins suggests that nongenetic factors play a major role in determining disease susceptibility. In this review, we consider how epigenetic mechanisms could affect the immune system and effector mechanisms in autoimmunity and/or the target organ of autoimmunity and thus affect the development of autoimmune diseases. We also consider the types of stimuli that lead to epigenetic modifications and how these relate to the epidemiology of autoimmune diseases and the biological pathways operative in different autoimmune diseases. Increasing our knowledge of these epigenetic mechanisms and processes will increase the prospects for controlling or preventing autoimmune diseases in the future through the use of drugs that target the epigenetic pathways.
Epigenetics and epigenetic mechanisms
What do we mean by “epigenetics”?
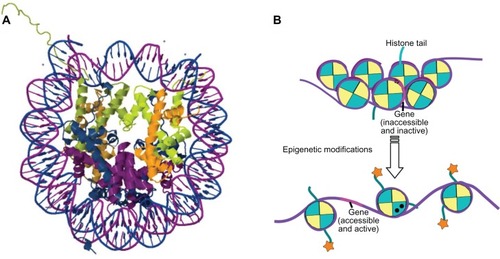
“Epigenetics” can be broadly defined as events or processes that affect the inheritance of gene activities but do not depend on any changes in DNA base sequences. The activity of genes is dependent largely on whether they are accessible to transcription factors; this is highly regulated by the dynamics of chromatin restructuring.Citation1 The basic repeating unit of chromatin is the nucleosome, in which ∼147 base pairs of negatively charged DNA wrap 1.65 times around a highly positively charged histone protein octamer, consisting of the H2A, H2B, H3, and H4 histone subunits (). The nucleosomes compact further to form the 30 nm fiber, with the H1 histone subunit playing an important role in stabilizing this structure. However, this tightly compacted chromatin structure poses barriers for processes such as transcription and replication, which require that the two strands of DNA come apart temporarily. Epigenetic “marks” on the chromatin play a central role in regulating the structure of chromatin () and thus the accessibility of DNA for transcription.Citation2
Figure 1 (A) Cartoon derived from the crystal structure (Protein Data Bank iD: 1aoiCitation298) of the histone octamer (H2A, blue; H2B, purple; H3, green; H4, orange) surrounded by 1.65 turns of DNA (∼147 base pair fragment). (B) In compacted chromatin, genes (represented in pink) are inaccessible and inactive, with hypermethylation of their promoter regions (▴), and there are few posttranslational modifications to the histones. Epigenetic modifications in the form of demethylation of gene promoter region, posttranslational modification of the histones (![]()
The actual term “epigenetics” was coined by CH Waddington in 1942 to describe the (at that stage, undefined) causal mechanisms by which the genes of a genotype could bring about phenotypic effects;Citation3 for example, the process by which differentiated cells maintain their specialized phenotypes through repeated cell division, with some genes remaining permanently switched on and others permanently switched off, even though all cells of an organism carry the same complement of DNA, was labeled an “epigenetic” control system.Citation4 It was not until the 1980s, however, that the term “epigenetics” was applied to a specific chemical modification, namely DNA methylation, that could affect the inheritance of gene activities without any changes in DNA base sequences.Citation5 Subsequently, the epigenetic label has also been applied to many histone modifications and other chromatin modifications that regulate transcription and/or replication,Citation6,Citation7 including gene silencing and X-inactivation, and to the regulatory actions of noncoding RNAs.Citation8 Some investigators have argued that histone modifications per se are not epigenetic,Citation9 as they have not been conclusively demonstrated to be self-propagating (heritable); however, as specific histone modifications can induce DNA methylation events,Citation10,Citation11 and DNA methylation events affect histone acetylation and histone methylation, there is an indirect heritability to these events.Citation12 More recently, several authors have attempted to clarify the definition of epigenetics to encompass all that it has come to mean;Citation13,Citation14 in this paper we will take a fairly broad definition of “epigenetics” as modifications that do not involve DNA base changes, that play a central role in controlling tissue and signal-specific gene expression, and that are responsible for the determination of gene expression profiles of tissues and cellular subsets.
Epigenetic modifications
Epigenetic modifications act by changing the way that DNA and histones interact in the nucleus, thereby allowing or preventing access by transcription factors and RNA polymerases, and regulating gene expression. Understanding the processes by which the epigenetic modifications result in a specific outcome is a very topical area of research, but there are still some large gaps in the specifics of how some of these modifications exert their effects.
DNA methylation
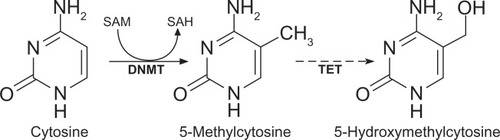
DNA methylation is an important epigenetic modification and is of great interest to autoimmunity, as treatment with the DNA methylation inhibitor 5-azacytidine is sufficient to induce autoimmune disease in experimental animals.Citation15,Citation16 In mammals, DNA methylation typically involves the attachment of a methyl group to cytosine moieties in CpG dinucleotides () and occurs at ∼70%–80% of CpG sites throughout the genome.Citation17 Methylated cytosine can be deaminated to thymine, either spontaneously or via enzymatic processes, leading to an underrepresentation (less than a quarter of what would be expected) of CpG sequences in the genome. However, dense clusters of short (∼500 bp) CpG-rich regions, known as “CpG islands,”Citation18,Citation19 also occur, particularly within the promoter region of genes, upstream from the transcriptional start sites (TSS), as many transcription factors require CpG-rich sites to bind to DNA.Citation20 The majority of CpG islands near TSS are hypomethylated; methylation of promoter CpG islands is usually linked to gene repression,Citation12 although it is still not clear if methylation precedes compaction of the nucleosomes, or whether it might act to “lock” the compacted nucleosomes.Citation21
Figure 2 Methylation of cytosines in DNA.
Abbreviation: SAH, S-adenosylhomocysteine.
Intragenic and intergenic CpG islands also occur and appear to be more susceptible to methylation than those in the promoter region.Citation22 It is thought that intragenic CpG islands may in some instances represent alternative TSS that could, for example, be utilized in a highly tissue-restricted fashion.Citation21,Citation23 Furthermore, methylation in the gene body can silence repetitive DNA elements, such as retroviral elements and, in contrast to methylation in promoter regions, also appears to actively stimulate transcription elongation.Citation19,Citation21 The functional significance of intergenic CpG islands is as yet unclear, although studies have localized at least some of these to promoters of noncoding RNAs,Citation24,Citation25 suggesting they may help regulate expression of these molecules.
DNA methylation also appears to play regulatory roles outside of CpG islands. For example, recent studies on colon cancer have described DNA hypo- or hypermethylation of oncogenes or tumor repressors, respectively, at “CpG island shores,” which lie up to 2 kb from CpG islands.Citation26 Recently, there have been several reports of regions of the genome that have only 10%–50% of the typical amount of CpG methylation and that appear to be involved in binding of transcription factors or nuclear receptors (such as the glucocorticoid receptor) in a cell-type specific manner.Citation27,Citation28 The formation of these “low-methylated regions” (LMRs) has been found to significantly correlate with increased expression of the nearest gene, whereas remethylation of LMRs coincides with reduced expression; this suggests that these LMRs represent distal regulatory regions.Citation27 Interestingly, 5-hydroxymethylcytosine (), which is produced by oxidation of 5-methylcytosine,Citation29 and TET1, which belongs to the family of ten-eleven translocation (TET) enzymes that catalyze this oxidative event,Citation30 appear to both be enriched in LMRs;Citation27 as 5-hydroxymethylcytosine has been reported to be present at enhancer regions,Citation31,Citation32 its enrichment in the LMRs provides support for the LMRs having a regulatory role.
Because DNA methylation plays such a critical role in regulating gene expression, the mechanisms for establishing, maintaining, and removing methyl groups have also been the subject of intense scrutiny, although there are still some major unanswered questions. DNA methylation of cytosines in mammalian cells occurs through the action of 5-methylcytosine DNA methyltransferases (DNMTs), which transfer a methyl group from a methyl group donor, S-adenosylmethionine, to the fifth carbon of the cytosine residues, converting them to 5-methylcytosines (). Five DNMTs have been identified in mammalian cells. It was thought for some time that DNMT1 could, by itself, maintain patterns of DNA methylation during cell replication; however, it is now recognized that ongoing participation from DNMT3a and DNMT3b is also required for methylation maintenance.Citation33 The latter are also critical for de novo methylation.Citation34
It has been reported that DNA methylation can be very dynamic, with some genes undergoing cyclical changes within minutes to hours,Citation35 but the mechanisms by which this type of rapid methylation/demethylation can occur are still not completely understood. Passive mechanisms of demethylation, such as occur when cells divide or when mammals erase genomic methylation patterns in primordial germ cells, are unlikely to account for such rapid cyclical changes in methylation patterns. However, the search for active demethylases has thus far not yielded very reproducible results.Citation36 It is thought that the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine via the action of TET family molecules may play a role in the more rapid cycles of methylation/demethylation, particularly in transcriptional regulatory regions such as enhancers.Citation21,Citation37
Initially, it was thought that methylation of CpG sites caused transcriptional repression by directly interfering with the binding of transcription factors to DNA;Citation38 however, it now appears that this is not the main mechanism by which methylated DNA inhibits gene expression. Instead, the methylated DNA attracts proteins that contain a methylated-DNA binding domain (MBD).Citation39,Citation40 MBD-containing proteins provide links between methylated DNA and covalent and non-covalent histone modifications that act to modify chromatin and repress gene activity.Citation41
Histone modifications and chromatin remodeling
Covalent modifications to the histone protein subunits can be made throughout the proteins; however, the N-termini of the histones, which extrude from the tightly wound chromatin strands, are particularly susceptible to posttranslational modifications. Such modifications can include acetylation, methylation, ubiquitylation, phosphorylation, and SUMOylation.Citation42 Acetylation/deacetylation and methylation/demethylation appear to be of most importance in regulating gene expression. Acetylation (via the action of histone acetyltransferases) of selected lysine residues in the tails of nucleosomal histones removes the positive charge on the lysine amino group that is acetylated, loosening the interaction between DNA and the histone and preventing chromatin from folding into the 30 nm fiber;Citation43 this typically promotes gene expression.Citation44,Citation45 In contrast, deacetylation (via histone deacetylases [HDACs]) is generally associated with gene repression. The phosphorylation state of histone H1 also appears to play a role in the folding of chromatin into higher-order structures.Citation46 The effects on gene regulation of methylation of lysine or arginine residues in the histone tails depend on both the specific site of the modified residues in the histone tail and the number of methyl groups that are added to these residues.Citation47,Citation48 In some cases, the same amino acid of a histone tail can be either acetylated or methylated with varying consequences; for example, lysine at position 9 of histone H3 (H3K9) is acetylated in regions of the genome that are transcriptionally active, but methylated in areas that are transcriptionally silent. Other specific histone modifications with well-defined effects on gene expression include H4 hyperacetylation and trimethylation of H3K4, both of which are present in many active genes.Citation49 Common sites of posttranslational histone modifications in H3 and H4 are shown in , although not all of these are necessarily epigenetic – some will probably be of more relevance to DNA replication and repair.
Table 1 Posttranslational modifications to tails of histones H3 and H4
Acetylation of lysine and methylation of lysine or arginine on histone tails plays a role in orchestrating the recruitment of multiprotein complexes. Some components of these complexes can “read” the histone changes via molecular interactions of the methyl/acetyl chains with structurally conserved protein modular domains that are present in many chromatin regulators and transcription factors and that act to change chromatin structure at target gene loci.Citation50 Some of these “readers” are extremely versatile: for example, proteins containing plant homeodomain (PHD) fingers appear to be able to read histone states in a sequence-dependent manner that is modulated positively or negatively by the methylation state of lysine and arginine, as well as by the acetylation state of lysine (reviewed in Sanchez and ZhouCitation51). Other components of the protein complexes recruited by the histone modifications are involved in chromatin remodeling: these components contain a conserved ATPase domain.Citation52 Remodeling utilizes energy from ATP hydrolysis to mobilize and restructure the nucleosomes and/or replace the histones themselves with histone variants.Citation52,Citation53 For example, remodeling complexes of the SWR1 family can replace H2A histones in H2A–H2B dimers with H2A.Z, forming variant nucleosomes with unique histone tails that can potentially bind unique regulatory proteins.Citation54 Other remodelers enable movement of nucleosomes along the DNA, thereby exposing new DNA to regulatory factors. Exactly how remodelers mobilize the nucleosome and how different remodeler complexes select which nucleosomes to move and restructure is still imperfectly understood.
Noncoding RNAs
Around 98% of the transcribed human genome does not encode proteinsCitation55 and recent studies have identified several different types of noncoding RNAs that act posttranscriptionally to regulate many biological processes via their interactions with messenger RNA (mRNA) or DNA.Citation16,Citation56,Citation57 Currently, those thought to be of most relevance to autoimmunity are the microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), although it is likely that further investigation will find that at least some of the other noncoding RNA types are also important.
The miRNAs are small molecules of around 22 nucleotides in length, which are believed to regulate the translation of more than 60% of protein-coding genes.Citation57,Citation58 In humans, more than 1000 miRNAs have been identified.Citation56,Citation59 Translation of mRNA into protein is blocked by human miRNAs primarily by inhibition of translation initiation either via binding of the miRNAs to the 3′UTRCitation57 or (less frequently in humans) via induction of mRNA degradation.Citation60 Some miRNAs regulate specific individual targets, while others appear to function as master regulators of a process, regulating expression levels of hundreds of genes simultaneously.Citation56,Citation57,Citation61 Recent studies suggest that there is strong bidirectional regulation between specific miRNAs and HDACs. For example, downregulation of miR-9 in bone marrow-derived CD19+ cells from patients with the B-cell lymphoma Waldenström’s macroglobulinemia results in upregulation of HDAC4 and HDAC5 expression levels and aberrant histone acetylation in these cells, and leads to abnormal expression of genes that cause the lymphoma.Citation62 Conversely, HDACs that are overexpressed in chronic lymphocytic leukemia block critical apoptosis-related miRNAs in the malignant B cells, resulting in pro-survival signals.Citation63
A fast-expanding area of research involves the role of lncRNAs in epigenetics. The definition of lncRNAs is complicated by the observation that lncRNAs often overlap with, or are interspersed between, multiple coding and noncoding transcripts; however, typically, they are more than 200 nucleotides in length and probably make up the largest portion of the mammalian noncoding transcriptome.Citation64 The lncRNAs appear to be able to recruit chromatin remodeling complexes to specific lociCitation65 and to regulate gene expression at the level of both transcriptionCitation66,Citation67 and posttranscriptional processing.Citation64,Citation68
Intersection of epigenetic pathways: X marks the spot
One of the best known processes where the various epigenetic pathways and mechanisms intersect is in the regulation of the X chromosome: normal females possess two X chromosomes and, in any given cell, one chromosome will be active (Xa) and one will be inactive (Xi),Citation69 whereas, in normal males, the X chromosome will remain active. However, during reproduction in mammals, the X chromosome undergoes its own cycle of inactivation and reactivation. The paternally derived X chromosome (XP) is imprinted in pre-implantation female embryos, leading to XP inactivation.Citation70 At implantation, the XP is reactivated and then, in the embryo proper (when there are ∼80–120 cells), X chromosome inactivation occurs randomly in each cell of the embryo,Citation71 so that all cells derived from each of these initial cells will maintain the same X-inactivation pattern. Thus, females are mosaics of two cell populations with respect to X-linked gene expression, one expressing the paternally derived genes and one expressing the maternally derived genes.
The X-inactivation center on each X chromosome contains the 17 kb lncRNA, X-inactivation specific transcript (Xist), which is necessary and sufficient for inactivation of the chromosome.Citation72,Citation73 Xist is involved in the transient imprinting of the XP during embryogenesis.Citation74,Citation75 In addition, during the inactivation process, the future Xi dramatically increases Xist RNA production and the Xist RNA spreads out from the X-inactivation center to progressively coat the chromosome;Citation73 in contrast, the future Xa ceases to express Xist. The silencing of genes along the Xi occurs soon after coating by Xist RNA; Xist appears to be crucial for initiating silencing but has a minor role in maintaining the Xi. Silence is maintained by packaging the Xi into densely packed, transcriptionally inactive heterochromatin, and is associated with high levels of DNA methylation, low levels of histone acetylation, low levels of histone H3K4 and H3R17 methylation, and high levels of histone H3K9 methylation.Citation76 Additionally, variants of histone H2A are exclusively found on nucleosomes along the Xi.Citation76 Thus, multiple layers of epigenetic modifications are involved in the process of X chromosome inactivation and silencing and they operate cooperatively to regulate gene expression. Given the predominance of autoimmunity in females,Citation77–Citation79 it is pertinent to question whether some of these mechanisms may fail in autoimmunity. This will be discussed in subsequent sections.
Studies of monozygotic (MZ) twins support a role for epigenetics in the development of autoimmunity
The etiology of the majority of autoimmune diseases remains obscure; however, for most, it appears that disease arises because of immune-, gene-, and environment-related effects. Evidence from studies of MZ twins shows that the penetrance of autoimmunity is typically in the order of only 20%–30% (), suggesting that a particular DNA nucleotide sequence alone is not sufficient for development of disease. For some of these diseases, widely varying concordance rates have been reported; it appears that factors such as the latitude of the country where the study participants livedCitation80,Citation81 and the age of disease onset in the probandCitation80 can influence the concordance rate, suggesting that environment and aging are major modifiers of the purely genetic effects. In addition, the number of years after diagnosis (in the proband) that the study was done appears also to be a determining factor in the concordance rate. For example, when MZ twin pairs initially discordant for type 1 diabetes (T1D) were followed over many years, the percentage of the initially unaffected twins who subsequently developed T1D increased at a slow rate until the age of 40, but then jumped from 25% at 40 years of age to over 60% at 60 years of age,Citation82 suggesting that, during the fifth or sixth decades of life, external and/or internal changes negate the protective effect that has prevented the initially healthy twin from developing disease up until that point. This would be most consistent with a role for epigenetic effects as modifiers of autoimmune disease penetrance, although clonal mosaicism that increases with age may also play a role.Citation83,Citation84
Table 2 Autoimmune diseases, showing female:male ratios of patients and concordance rates in monozygotic (MZ) twins
To investigate the role of epigenetics, several studies have looked at whether there are differences in DNA methylation in MZ twins discordant for autoimmune disease. The evidence is variable. Javierre et alCitation85 found DNA methylation changes in MZ twins discordant for systemic lupus erythematosus (SLE), but not for rheumatoid arthritis (RA) or dermatomyositis. In studies of female twin pairs discordant for multiple sclerosis (MS), Baranzini et alCitation86 found few differences between female MZ twin pairs with respect to either DNA sequences, DNA methylation, or RNA sequences; however, because only extreme methylation differences (a change of at least 60%) were investigated in this study, and only far smaller differences have been identified in some other studies using nonmalignant tissues,Citation87–Citation89 potential variation could have been missed. In MZ twin pairs discordant for psoriasis, differences in DNA methylation between unaffected and affected twins were correlated with differences in gene expression, particularly in CD4+ T cells.Citation90 A recent study looked for methylation differences in CD14+ monocytes from MZ twin pairs discordant for T1D and identified 132 different CpG sites at which the direction of the intra-MZ pair DNA methylation difference significantly correlated with the diabetic state.Citation91 Of course, DNA methylation is not the only epigenetic change that might explain the lack of concordance in MZ twin pairs, but as yet there are few studies investigating other potential mechanisms. Furthermore, most of these studies only investigated small numbers of twin pairs and most used peripheral blood containing multiple cell types (and therefore multiple epigenomes) as the source of DNA, which could skew methylation differences,Citation92 making it difficult to draw global conclusions from the results.
Evidence for epigenetic modifications in autoimmune diseases
Each autoimmune disease involves at least two major players, the immune system, which is the effector of the autoimmune damage, and the target organ, which can have variable degrees of resistance to autoimmune damage. Epigenetic modifications to either the immune system or the target organ could play a role in disease development. In the following section, we will review some of the modifications that have been found thus far, but major questions remain unanswered, such as: Are the same epigenetic modifications seen in all patients with the same clinical phenotype? How do the epigenetic effects interact with polymorphisms in autoimmune susceptibility genes? Is there a threshold in the number/type of epigenetic modifications that need to occur for disease to become manifest?
Epigenetic modifications in the immune system
Numerous recent studies clearly indicate that there is epigenetic control of major immune cell functions, such as hematopoietic lineage choice, antigen-receptor rearrangement and allelic exclusion, and immune responses to pathogens.Citation93–Citation105 For example: epigenetic processes govern the differentiation of T helper cells and their lineage stability,Citation93,Citation105 exposure of dendritic cells to oxidized phospholipids can change the phenotype of the cells through epigenetic mechanisms,Citation106 and epigenetic processes control the production of antibodies by B cells.Citation107 These immunological developmental events that are regulated by epigenetic mechanisms could possibly be altered to promote autoimmunity. In this section, we will focus on some of the epigenetic modifications that could specifically affect susceptibility to, or development of, autoimmune diseases.
Epigenetic control of immune tolerance
One of the major mechanisms in the control of development of autoreactive immune cells is induction of T cell central tolerance in the thymus. Central tolerance against many autoantigens is regulated by the autoimmune regulator (AIRE) protein, which promotes expression of tissue-specific antigens in thymic medullary epithelial cells. AIRE contains PHD fingers, which bind to methylated histone H3 and provide a link between the status of histone modifications and the regulation of tissue-specific antigen expression in the thymus. In an animal model with a targeted mutation within one of these PHD fingers, there was decreased binding of AIRE to methylated histone H3, a generalized dampening of AIRE’s transcriptional impact, and development of autoimmunity.Citation108
Another potential mechanism by which epigenetic mechanisms could affect central tolerance is via the action of the lncRNA, growth-arrest-specific 5 (GAS5). GAS5 sensitizes cells to apoptosis by regulating the activity of glucocorticoids, is present in T cells,Citation109 and has been linked to increased susceptibility to autoimmune disorders, including SLE.Citation110 Positive selection of T cells in the thymus is postulated to be dependent on the counter-interaction between glucocorticoid receptor- and T cell receptor-induced death signals.Citation111 Modulation of GAS5 expression levels could regulate selection of T cells through the glucocorticoid receptor, thereby providing resistance to apoptosis, allowing escape of self-reactive T cells, and potentiating development of autoimmunity.
There is evidence that miRNAs also play a major role in induction of central tolerance. For example, the affinity of maturing T cells for antigen during thymic development plays a large role in determining their fate: there appears to be a strong correlation between the sensitivity of the T cells to antigen and levels of miR-181a, which is dynamically regulated during T cell maturation in the thymus.Citation112 In addition, recent studies have identified a selection of miRNAs that have known functions in thymopoiesis and which are differentially regulated in the thymus following stress; this dysregulation has the potential to alter T cell repertoire selection and the formation of naive T cells.Citation113
In the periphery, some of the best-studied mediators of tolerance are the CD4+ Foxp3+ regulatory T cells (Tregs). Foxp3 is known to form complexes with histone acetyltransferases (Tip60 and p300) or HDACs (HDAC7); these complexes then epigenetically modulate target gene expression via histone or Foxp3 acetylation or deacetylation.Citation114,Citation115 Recently, it was shown that mice in which Foxp3 failed to associate with Tip60 and HDAC7 had altered Foxp3-dependent transcription and epigenetic modifications, causing insufficiency in natural Tregs and impaired development of inducible Tregs.Citation116 It has been reported that Foxp3 also represses the activity of SATB1, a genome organizer that regulates chromatin structure and gene expression, leading to loss of suppressive function in the Tregs and establishment of effector functions.Citation117 Foxp3 not only acted directly as a transcriptional repressor of SATB1 but also indirectly through the induction of miRNAs that bind the SATB1 3′ untranslated region. Other studies have also highlighted the role that miR-155 plays in Treg development and that miR-146 and the miR-17-92 cluster play in Treg function (as recently reviewed in Zhou et al, 2011).Citation118
Other cells involved in maintenance of peripheral tolerance include natural killer T cells and regulatory subsets of dendritic cells. Several reports of miRNA- or HDAC-mediated regulation of these subsets of cells exist, although most have not specifically addressed the issue of how levels of these molecules change when peripheral tolerance is compromised.Citation118,Citation119 Another study has suggested that programmed death-1, a negative regulatory molecule expressed on activated CD4+ and CD8+ T cells, maintains immune homeostasis and self-tolerance through a miRNA (miR-21) signaling cascade.Citation120
The studies looking at epigenetic control of tolerance have probably only touched the tip of the iceberg with respect to the complexity of the levels of control and likely mechanisms; however, they do provide support that this is one way in which epigenetics could affect autoimmunity.
Epigenetics and human leukocyte antigen (HLA)-disease associations
A large number of autoimmune diseases are strongly linked to carriage of specific HLA (particularly HLA class II) molecules; interestingly, however, HLA linkage for some autoimmune diseases varies around the globe. In MS, for example, linkage is with DRB1*1501 in Caucasian populations and DRB1*0301, DRB1*0405, and DRB1*1303 in Sardinia, but DRB1*07 in continental Italy and DRB1*13 in Israel. It has been shown that coordinated changes of histone modifications and HDAC mobilization regulate the induction of HLA class II genesCitation121 and proposed that the different HLA associations observed in MS patients across the globe are a reflection of specific environmental factors influencing epigenetic marks on liable haplotypes, which affect the expression or function of class II genes and permit the MS pathogenic cascade.Citation122 In MS, there is also some evidence of epigenetic modification of HLA molecules contributing to the inheritance of disease susceptibility, particularly transmission from mothers to offspring.Citation123,Citation124
Other diseases have been less well studied, but in autoimmune diseases where HLA linkage is important, epigenetic modification of the HLA molecules could possibly modify the HLA linkage. In diseases such as ankylosing spondylitis, where there is strong HLA linkage and good concordance between MZ twins, this is less likely to play a role.
Abnormal epigenetic marks on peripheral blood mononuclear cell (PBMC) subsets
There have been multiple reports of altered levels of epigenetic marks in PBMCs and subsets of these, particularly B and T cells, from patients with autoimmune disease compared with healthy controls: these are summarized in . Some alterations appear to be relatively disease specific, whereas others may relate more to the autoimmune phenotype. PBMCs from patients with SLE have been studied extensively and recently reviewed:Citation125,Citation126 in brief, there is global hypomethylation of B and T cells, alterations in DNMT expression and in histone acetylation, particularly affecting genes related to apoptosis, and changes to levels of specific miRNAs. The mechanisms by which these changes in epigenetic marks modulate specific cellular processes are currently the subject of intense research interest.
Table 3 Abnormal epigenetic marks on peripheral blood leukocytes in autoimmune diseases
Epigenetic modifications in the target organ
As noted, epigenetic changes in some autoimmune diseases are particularly notable in cells of the immune system; however, changes in the target organ could also render the organ more susceptible to autoimmune attack. Such failure of target-organ resistance could play a role in the development of autoimmune disease.Citation127,Citation128 What is still unknown is whether epigenetic changes in both the immune system and the target organ are required for development of autoimmune disease, whether changes in one are more likely to occur than changes in the other (eg, whether organs/cell systems that turn over more rapidly are more likely to be affected), or whether the type of autoimmune disease (ie, organ specific vs systemic) correlates more with epigenetic changes in the target organ than in the immune system.
During the last 2–3 years, a substantial body of evidence has accumulated to suggest that epigenetic changes in the target organ are important in some autoimmune diseases. For example, in synovial tissues from patients with RA, there is hypomethylation of DNA,Citation129 decreases in levels of HDAC1 and HDAC2 and hyperacetylation of histones H3 and H4,Citation130 and hypomethylation of histone H3 at lysine 9. Similarly, in MS patients, compared with healthy individuals, there is hypomethylation of DNA from normal-appearing central nervous system white matter, but not from tissue in the thymus, demonstrating that the target organ alone can show epigenetic changes.Citation131 In anti-neutrophil cytoplasmic antibody vasculitis, there is upregulation of the target antigen due to perturbation of epigenetic gene-silencing mechanisms.Citation132 SLE is a special case when considering the target organ, as the target organ of SLE, the cell nucleus, is also the site of the epigenetic effects. Multiple studies over a number of years have shown that modifications to both DNA and histones can affect their antigenicity. Apoptotic hypomethylated DNA and posttranslationally modified histones are known to be major targets of autoantibodies in SLE.Citation133–Citation136 lists changes in epigenetic marks that are seen in the target tissue of some autoimmune diseases. In some cases, the interpretation of the target-tissue data might be made more difficult by the presence of immune cells or other inflammatory mediators within the target organ.
Table 4 Changes in epigenetic marks seen in the target tissue of some autoimmune diseases
What stimuli might induce epigenetic changes that lead to the development of autoimmunity?
In most cases, the nature of the specific stimuli that lead to the epigenetic changes seen in patients with autoimmune disease remain undefined, but could include many diverse stimuli, both external (eg, diet, exposure to sunlight, environmental chemicals, toxins, and drugs/pharmaceuticals) and internal (eg, aging, stress, sex hormones). A complicating factor in studies of stimuli of epigenetic changes is that, while some epigenetic effects are manifested in the generation of patients that are exposed to the modifying agent, in other cases, it appears that disease occurs predominantly one or two generations after the exposure.Citation137
It is also of interest to consider how different stimuli might differentially affect males and females, given the higher incidence of autoimmunity in females ().Citation77–Citation79 For epigenetic modifications to explain sexual dimorphism in autoimmunity, it is necessary to suppose that epigenetic factors are more commonly encountered in one sex than in the other, or that members of one sex are more vulnerable than the other: this is speculative. However, in view of the increase in some autoimmune diseases among women, but not men, over the last 100 years,Citation138–Citation142 and the major societal changes that have particularly affected women over that time, it is an attractive hypothesis.
Effects of external exposure to agents that induce epigenetic changes
Diet and nutrition
The diet provides the methyl donors (methionine, choline) and cofactors (folic acid, vitamin B12 and pyridoxal phosphate) essential for DNA and histone methylation.Citation143 It is now well recognized that susceptibility to adult-onset chronic disease is influenced by persistent adaptations to prenatal and early postnatal nutrition.Citation144,Citation145 Furthermore, there are also reports of diet-induced epigenetic changes in the adult state. For example, dietary components such as alcohol, vitamin B6, and vitamin A have been linked to DNA methylation within the gastrointestinal tract and development of colorectal cancerCitation146–Citation149 and diets such as the “epigenetic diet” have been popularized as a means of controlling cancer via epigenetic modification.Citation150
It has been proposed that epigenetic links between nutrition and autoimmunity may well contribute to the epidemiology observed for numerous autoimmune diseases;Citation151 however, while studies in animals using arbitrarily chosen dietary elements tend to support this proposal, human data from real-life clinical settings or randomized clinical trials remains inconclusive at present. There are, however, suggestions that obesity may predispose to autoimmune disease.Citation152 In MS, for example, either maternal obesity and diabetesCitation153 or teenage obesityCitation154 may predispose to later MS; increased lipid levels are associated with worse outcome;Citation155 and increased levels of the adipose-derived hormone, leptin, are associated with decreased numbers of Tregs in patients with MS.Citation156 There is much evidence to support epigenetic regulation of numerous genes, including many immune-related genes, as a consequence of obesity,Citation157 although such studies are thus far lacking in obese individuals with autoimmune disease.
Environmental exposures
There is considerable evidence that prevalence rates for autoimmune diseases, including MS, T1D, RA, dermatomyositis, and polymyositis,Citation158–Citation161 increase as latitude increases; thus, it is thought that increased exposure to sunlight and ultraviolet radiation (UVR) in areas of lower latitude might be protective, possibly via vitamin D,Citation162 although recent studies suggest that the protective effects of UVR are not necessarily dependent on vitamin D.Citation163 UVR is known to induce multiple epigenetic changes, many of which are immunosuppressive, including hypermethylation of numerous gene promoters, phosphorylation of histone H3, and acetylation of histones H3 and H4.Citation164,Citation165 A possible explanation for reduced autoimmunity in men compared with women would be that, generally, males have more sun exposure and use less sun protection than females;Citation166 however, while females may have less UVR exposure, there could also be biological differences between males and females in response to UVR, as there are gender differences in the degree of carcinogenesis and inflammation after ultraviolet exposure in mice.Citation167 With respect to vitamin D, it is now known that vitamin D controls the transcription of many genes through multiple epigenetic effects via vitamin D response elements.Citation168 In addition, there appear to be gender differences in the effects of vitamin D on gene expression;Citation169 it has been shown, for example, that higher levels of vitamin D are associated with a lower incidence of MS only in women.Citation170
There are many reports of “outbreaks” of autoimmunity, which tend to occur in areas where there is increased exposure to heavy metals.Citation171,Citation172 It has been found that nickel, cadmium, lead, aluminum, mercury, and arsenic (all of which are widespread environmental contaminants) can exert immunomodulatory and toxic effects through epigenetic mechanisms such as global changes to DNA methylation, histone modifications, and changes in levels of miRNAs.Citation173,Citation174
Another common environmental exposure that has been linked to increased prevalence of many autoimmune diseases is tobacco smoke.Citation175 In addition to nicotine, smokers are exposed to over 6000 other chemicals generated by the burning tobacco, many of which are known to be antigenic, cytotoxic, mutagenic, or carcinogenic. A recent study suggests that maternal smoking can deregulate placental methylation in a CpG site-specific manner that correlates with meaningful alterations in gene expression, particularly along oxidative stress pathways.Citation176
Drugs/pharmaceuticals
Several drugs, most notably procainamide and hydralazine, are known to cause increases in antinuclear antibodies in most people and lupus-like symptoms in some individuals.Citation177 Both procainamide and hydralazine induce hypomethylation of DNA, but via different pathways: procainamide is a competitive inhibitor of DNMT1Citation178 whereas hydralazine inhibits DNMT1 upregulation during mitosis by blocking the extracellular signal-regulated kinase (ERK) signaling pathway at PKCδ.Citation179 Chemicals in cosmetics have also been suggested to act as a trigger for primary biliary cirrhosis,Citation180 but the exact mechanisms by which these might act have not yet been elucidated, and thus far there are no studies regarding cosmetics and other autoimmune diseases.
Chemicals that mimic the effects of estrogen (eg, phytoestrogens or drugs such as diethylstilbestrol) are known to exert major epigenetic effects during fetal development,Citation181–Citation183 with the dose and timing of exposure in relation to the developmental age of the fetus markedly affecting the outcomes. Thus far, evidence linking these to autoimmune disease is equivocal,Citation184 although studies are limited. Another possibility that has been considered is that exposure to the oral contraceptive pill might have influenced increasing female incidence of autoimmune disease over the last 30–40 years. Recent studies show that females using the oral contraceptive pill have lower global DNA methylation levels than females who do not use oral contraceptives;Citation185 however, no specific link between the incidence of autoimmune disease and use of oral contraceptives has yet been identified. There have been reports of worsening of autoimmune disease following assisted reproduction treatment (ART),Citation186 but so far no clear evidence that children conceived through ART have an increased risk of developing autoimmunity (although the oldest of these children are only now in their early thirties). ART generally involves the use of hormone therapy (with gonadotropin-releasing hormone agonists, follicle-stimulating hormone, luteinizing hormone, human chorionic gonadotropin, or progesterone) to induce ovulation or to assist in implantation. Recent studies suggest that ART is associated with lower mean methylation at CpG sites in the placenta and higher mean methylation at CpG sites in cord blood, on both imprinted and non-imprinted genes;Citation187,Citation188 whether or not these observations will translate to increased levels of autoimmunity in children conceived in this manner remains to be seen. No studies have specifically addressed the issue of whether ART might induce detrimental epigenetic changes in mothers who already have an autoimmune disease.
Retroviruses
Human retroviruses have been linked to multiple autoimmune diseases for many years,Citation189 although absolute proof that they cause disease is still lacking. However, a large part (∼8%) of the human genome is comprised of human endogenous retroviral elements (HERVs) that have incorporated into the primate genome over millions of years. Normally, HERVs are repressed by DNA methylation and other mechanisms; however, it has recently been reported that there is hypomethylation of the HERV long interspersed repetitive element 1 in B and T cells of SLE patients.Citation190 This could lead to expression of retroviral proteins in the lymphocytes, with potential consequences for the integrity, physiology, and immune function of these cells and subsequent development of autoimmunity.
“Internal” factors that could lead to epigenetic changes of relevance to autoimmunity
Aging
Aging is associated with changes in patterns of gene expression; whether these changes are due primarily to epigenetic alterations or genetic alterations (ie, accumulated DNA damage) is not clear, but it is feasible that epigenetics accounts for at least part of the change. Early studies in mammals found age-associated increases in methylation of CpG islandsCitation191 and of histone H4K20;Citation192 both of these epigenetic marks correlate with transcriptional repression. More recent studies of twins have shown that, although MZ twins are epigenetically indistinguishable during the early years of life, older MZ twins exhibit many differences in their overall content and genomic distribution of methylated DNA and histone acetylation and their gene-expression portrait.Citation193,Citation194 In a genome-scale study of epigenomic dynamics during normal human aging, Rakyan et alCitation195 identified aging-associated differentially methylated regions, which became hypermethylated with aging, predominantly at bivalent chromatin domain promoters. Bivalent chromatin domains, which contain two methylation sites with conflicting output (H3K4 and H327 – see ) are characteristic of pluripotent cells, but methylation of one site dominates as the cells differentiate.Citation196 This is of interest, as Polycomb-group proteins catalyze the methylation of H3K27, and it has also been reported that Polycomb-group protein target genes are far more likely to become methylated with age than nontargets.Citation197 Since most autoimmune diseases do not develop until the third or fourth decade of life (with notable exceptions such as T1D), accumulation of epigenetic changes with aging may provide the impetus for the onset of disease in susceptible individuals.
Stress
Physical and psychological stresses have been suggested as being potential modulators in the development of autoimmune disease.Citation198 Hypomethylation of promoter regions of genes encoding glucocorticoid receptors and brain-derived neurotrophic factor have been reported fairly frequently in offspring whose mothers experienced stressful events during pregnancy. Adolescents whose parents experienced adversity during the adolescent’s infancy or preschool years also showed differences in the numbers of methylated CpG sites compared with controls, although these differences were not found to be associated with specific genes.Citation199 Interestingly, maternal stressors in infancy, but paternal stressors in the preschool years, are most strongly predictive of differential methylation and the patterning of such epigenetic marks varies by the child’s gender, with girls showing higher levels of differentially methylated CpG sites than boys.Citation199
Quality of early maternal care has also long been acknowledged to have long-term repercussions during the lifetime of an individual. Studies in rats have shown that, in the absence of appropriate maternal nurturing, there is less methylation of the gene encoding the glucocorticoid receptor in the hippocampus, resulting in overexpression of the receptor in later life.Citation200 The implication is that the glucocorticoid-mediated stress-response pathway is epigenetically fixed at the level of gene transcription.
Effects of exposure to estrogen
As noted earlier, chemicals that can mimic the effects of estrogen can exert major epigenetic effects during fetal development; similarly, levels of naturally occurring estrogens in the mother during pregnancy can also epigenetically influence fetal development, particularly in the developing brain.Citation201 Almost all of the papers looking at the epigenetic modifying effects of estrogens consider them in terms of effects on the fetus; surprisingly little work has been reported regarding the epigenetic effects on the mother of the greatly elevated levels of estrogens during pregnancy.Citation202 However, the state of being pregnant has short- and long-term beneficial effects on some females with autoimmune disease. In MS, for example, there are reports of favorable long-term effects of childbirth on the course of MS, particularly if the child was born after the onset of MS,Citation203–Citation205 and women who have never had children are reported to take a shorter time to reach a higher level of disability than are women who have had children at any time.Citation203 While these studies could simply indicate that women with more aggressive disease are just less inclined to have children, another possibility is that pregnancy induces long-term protective epigenetic effects in the mother. To understand how/if epigenetic effects of estrogen or other pregnancy-related hormones might help to improve autoimmunity, it will first be necessary to determine the epigenetic marks associated with these hormones in normal healthy cells and organs and compare these with the same patterns in diseased or dysfunctional ones. This information is still lacking.
Exercise
Exercise has numerous health benefits and has effects on the immune system, where it has been shown that exercise in low levels is anti-inflammatory.Citation206 Anecdotal evidence suggests a beneficial role of exercise in autoimmunity, although data from controlled trials is scarce.Citation207 It has been reported that exercise can alter gene expression in skeletal muscle through epigenetic mechanisms, including gene-specific DNA hypomethylation,Citation208 and increased acetylation of H3K36, a site associated with transcriptional elongation, combined with export from the nucleus of HDAC4 and HDAC5 (thereby removing their transcriptional repressive function).Citation209 Although these observations are from small numbers of individuals, and some of the effects reported are relatively small, they suggest that at least some of the effects of exercise may come about via epigenetic modifications. If this were confirmed, it would be plausible that many of the effects of exercise in other tissues, including the immune system, also occur through epigenetic modifications.
Epigenetic modification to the X chromosome
X-linked genes are typically unmethylated (active) in men, while women have one methylated (Xi) and one unmethylated (Xa) gene. It has been found that some genes on Xi are demethylated in females with autoimmune disease; this has been studied most thoroughly in SLE.Citation210 The consequence of this appears to be that females can have elevated levels of expression of molecules encoded on the X chromosome, due to gene dosage irregularities. In support of this, it has been shown that treatment of CD4+ T cells with the DNMT inhibitor 5-azacytidine leads to elevated levels of CD40 ligand (CD154 – which is encoded on the X chromosome) on T cells from females but not males.Citation211 There are observations of elevated levels of CD154 in a multitude of autoimmune disorders, including SLE, autoimmune thyroid disease, T1D, inflammatory bowel disease, psoriasis, MS, and RA. Although the gender of patients has not always been specified in these studies, the results suggest that demethylation of genes on the Xi may be a fairly common epigenetic mechanism in autoimmune disease. Several molecules thought to be of great potential importance for autoimmunity are encoded on the X chromosome; these include: the interleukin (IL) 2Rγ chain (also known as the “common γ chain” because it is shared by receptors for IL2, IL7, IL15, and IL21);Citation212 Foxp3, the master regulator in the development and function of Tregs;Citation213 tissue inhibitors of metalloproteinases 1–4, which are important in inflammation;Citation214 the X-linked inhibitor of apoptosis that regulates T cell function;Citation215 and toll-like receptor 7 and toll-like receptor 8, which are pattern-recognition receptors, both of which are important in recognition of single-stranded RNA.Citation216 There are also a large number of X-linked genes encoding possible target antigens of autoimmune disease: changes in dosage of these autoantigens could also provide an initiating event for development of autoimmunity. This is an area of research that is just opening up and it is likely that the next few years will dramatically increase our understanding of how X chromosome regulation might be involved in the development of autoimmunity.
Skewing of X inactivation, so that there is an overrepresentation of the Xa from one parent, has been reported in females with some autoimmune diseases, including autoimmune thyroid disease and scleroderma,Citation217–Citation220 but not in SLE or MS.Citation221,Citation222 As noted earlier, the process of X inactivation has multiple layers of epigenetic controls; thus, the potential for modulation of this process is high. Skewed X inactivation could influence expression of genes on the X chromosome and could lead to inactivation of a gene that protects against autoimmunity, or overexpression of a susceptibility gene, leading to increased autoimmune disease.
The next steps
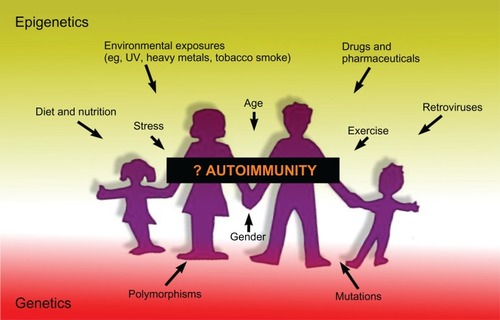
Experimental evidence of a major role for epigenetically modified gene expression in the development of autoimmunity is accumulating at an ever-increasing pace, but there are many aspects still not understood. Over the last couple of years, the global methylation measures, which have been used in many of the studies noted in this review, have been superseded by new technologies involving next-generation sequencing that can dramatically increase the amount and detail of the information that can be obtained regarding epigenetic modifications.Citation223–Citation226 Integrating the extensive genome-wide association study data that are available from many autoimmune diseases with methylome, chromatin epigenomic, and ncRNA data would be one way to potentially maximize our understanding of how the interaction of genetic and epigenetic factors increases susceptibility to or protection against autoimmune disease (). Although, as has been pointed out,Citation227 because epigenetic marks are known to change over the lifetime of an individual, it is harder to study with the same certainty as genetic association the association of epigenetics with disease.
Figure 3 Summary of factors that may lead to the development of autoimmunity.
Abbreviation: UV, ultraviolet.
However, improving our understanding of the role that epigenetic modifications play in the development of autoimmunity is likely to increase the prospects for controlling or preventing autoimmune disease through the use of drugs that target proteins controlling chromatin modifications (eg, HDAC inhibitors), DNA methylation (eg, inhibitors of DNA methyltransferases), or other epigenetic mechanisms.
Disclosure
The authors declare no conflicts of interest in this work.
References
- LiBCareyMWorkmanJLThe role of chromatin during transcriptionCell2007128470771917320508
- MargueronRReinbergDChromatin structure and the inheritance of epigenetic informationNat Rev Genet201011428529620300089
- WaddingtonCHThe epigenotypeInt J Epidemiol2012411101322186258
- NanneyDLEpigenetic control systemsProc Natl Acad Sci U S A195844771271716590265
- HollidayRThe inheritance of epigenetic defectsScience198723848241631703310230
- JeppesenPHistone acetylation: a possible mechanism for the inheritance of cell memory at mitosisBioessays199719167749008418
- TurnerBMHistone acetylation and an epigenetic codeBioessays200022983684510944586
- GrewalSIMoazedDHeterochromatin and epigenetic control of gene expressionScience2003301563479880212907790
- PtashneMOn the use of the word ‘epigenetic’Curr Biol2007177R233R23617407749
- OoiSKQiuCBernsteinEDNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNANature2007448715471471717687327
- ZhangYReinbergDTranscription regulation by histone methylation: interplay between different covalent modifications of the core histone tailsGene Dev200115182343236011562345
- KloseRJBirdAPGenomic DNA methylation: the mark and its mediatorsTrends Biochem Sci2006312899716403636
- BirdAPerceptions of epigeneticsNature2007447714339639817522671
- HaigDCommentary: The epidemiology of epigeneticsInt J Epidemiol2012411131622186259
- HewagamaARichardsonBThe genetics and epigenetics of autoimmune diseasesJ Autoimmun200933131119349147
- MedaFFolciMBaccarelliASelmiCThe epigenetics of autoimmunityCell Mol Immunol20118322623621278766
- EhrlichMGama-SosaMAHuangLHAmount and distribution of 5-methylcytosine in human DNA from different types of tissues of cellsNucleic Acids Res1982108270927217079182
- TakaiDJonesPAComprehensive analysis of CpG islands in human chromosomes 21 and 22Proc Natl Acad Sci U S A20029963740374511891299
- IllingworthRSBirdAPCpG islands – ‘a rough guide’FEBS Lett2009583111713172019376112
- Mariño-RamírezLSpougeJLKangaGCLandsmanDStatistical analysis of over-represented words in human promoter sequencesNucleic Acids Res200432394995814963262
- JonesPAFunctions of DNA methylation: islands, start sites, gene bodies and beyondNat Rev Genet201213748449222641018
- IllingworthRKerrADeSousaDA novel CpG island set identifies tissue-specific methylation at developmental gene lociPLoS Biol200861e2218232738
- MaunakeaAKNagarajanRPBilenkyMConserved role of intragenic DNA methylation in regulating alternative promotersNature2010466730325325720613842
- PanningBJaenischRDNA hypomethylation can activate Xist expression and silence X-linked genesGene Dev19961016199120028769643
- SleutelsFZwartRBarlowDPThe non-coding Air RNA is required for silencing autosomal imprinted genesNature2002415687381081311845212
- IrizarryRALadd-AcostaCWenBThe human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shoresNat Genet200941217818619151715
- StadlerMBMurrRBurgerLDNA-binding factors shape the mouse methylome at distal regulatory regionsNature2011480737849049522170606
- WienchMJohnSBaekSDNA methylation status predicts cell type-specific enhancer activityEMBO J201130153028303921701563
- KriaucionisSHeintzNThe nuclear DNA base 5-Hydroxymethylcytosine is present in Purkinje neurons and the brainScience2009324592992993019372393
- WilliamsKChristensenJHelinKDNA methylation: TET proteins – guardians of CpG islands?EMBO Rep2012131283522157888
- PastorWAPapeUJHuangYGenome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cellsNature2011473734739439721552279
- StroudHFengSMorey KinneySPradhanSJacobsenSE5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cellsGenome Biol2011126R5421689397
- JonesPALiangGRethinking how DNA methylation patterns are maintainedNat Rev Genet2009101180581119789556
- BallestarEAn introduction to epigeneticsAdv Exp Med Biol201171111121627038
- MétivierRGallaisRTiffocheCCyclical DNA methylation of a transcriptionally active promoterNature20084527183455018322525
- OoiSKBestorTHThe colorful history of active DNA demethylationCell200813371145114818585349
- BoothMJBrancoMRFiczGQuantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolutionScience2012336608393493722539555
- EdenSCedarHRole of DNA methylation in the regulation of transcriptionCurr Opin Genet Dev1994422552598032203
- CurradiMIzzoABadaraccoGLandsbergerNMolecular mechanisms of gene silencing mediated by DNA methylationMol Cell Biol20022293157317311940673
- HendrichBBirdAIdentification and characterization of a family of mammalian methyl-CpG binding proteinsMol Cell Biol19981811653865479774669
- SmithCLPetersonCLA conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodelingMol Cell Biol200525145880589215988005
- FischleWTsengBSDormannHLRegulation of HP1-chromatin binding by histone H3 methylation and phosphorylationNature200543870711116112216222246
- Shogren-KnaakMIshiiHSunJMPazinMJDavieJRPetersonCLHistone H4-K16 acetylation controls chromatin structure and protein interactionsScience2006311576284484716469925
- GregoryPDWagnerKHörzWHistone acetylation and chromatin remodelingExp Cell Res2001265219520211302684
- NeumannHHancockSMBuningRA method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylationMol Cell200936115316319818718
- LiGMargueronRHuGStokesDWangYHReinbergDHighly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivoMol Cell2010381415320385088
- ChenPLiGDynamics of the higher-order structure of chromatinProtein Cell201011196797121153512
- ScharfANImhofAEvery methyl counts – epigenetic calculusFEBS Lett2011585132001200721108946
- BallestarEEpigenetic alterations in autoimmune rheumatic diseasesNat Rev Rheumatol20117526327121343899
- KouzaridesTChromatin modifications and their functionCell2007128469370517320507
- SanchezRZhouMMThe PHD finger: a versatile epigenome readerTrends Biochem Sci201136736437221514168
- ClapierCRCairnsBRThe biology of chromatin remodeling complexesAnn Rev Biochem20097827330419355820
- MinardMEJainAKBartonMCAnalysis of epigenetic alterations to chromatin during developmentGenesis200947855957219603511
- MizuguchiGShenXLandryJWuWHSenSWuCATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complexScience2004303565634334814645854
- MattickJSChallenging the dogma: the hidden layer of non-protein-coding RNAs in complex organismsBioessays2003251093093914505360
- EstellerMNon-coding RNAs in human diseaseNat Rev Genet2011121286187422094949
- HeLHannonGJMicroRNAs: small RNAs with a big role in gene regulationNat Rev Genet20045752253115211354
- FriedmanRCFarhKKBurgeCBBartelDPMost mammalian mRNAs are conserved targets of microRNAsGenome Res20091919210518955434
- BentwichIAvnielAKarovYIdentification of hundreds of conserved and nonconserved human microRNAsNat Genet200537776677015965474
- YektaSShihIBartelDPMicroRNA-directed cleavage of HOXB8 mRNAScience2004304567059459615105502
- MendellJTMicroRNAs: critical regulators of development, cellular physiology and malignancyCell Cycle2005491179118416096373
- RoccaroAMSaccoAJiaXmicroRNA-dependent modulation of histone acetylation in Waldenström macroglobulinemiaBlood201011691506151420519629
- SampathDLiuCVasanKHistone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemiaBlood201211951162117222096249
- MercerTRDingerMEMattickJSLong non-coding RNAs: insights into functionsNat Rev Genet200910315515919188922
- GuptaRAShahNWangKCLong non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasisNature201046472911071107620393566
- WangXAraiSSongXInduced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcriptionNature2008454720012613018509338
- MartianovIRamadassASerra BarrosAChowNAkoulitchevARepression of the human dihydrofolate reductase gene by a non-coding interfering transcriptNature2007445712866667017237763
- HeYVogelsteinBVelculescuVEPapadopoulosNKinzlerKWThe antisense transcriptomes of human cellsScience200832259091855185719056939
- LyonMFGene action in the X-chromosome of the mouse (Mus musculus L.)Nature196119037237313764598
- TakagiNSasakiMPreferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouseNature197525655196406421152998
- RastanSTiming of X-chromosome inactivation in postimplantation mouse embryosJ Embryol Exp Morphol19827111246185603
- PennyGDKayGFSheardownSARastanSBrockdorffNRequirement for Xist in X chromosome inactivationNature199637965611311378538762
- HerzingLBRomerJTHornJMAshworthAXist has properties of the X-chromosome inactivation centreNature199738666222722759069284
- OkamotoIOtteAPAllisCDReinbergDHeardEEpigenetic dynamics of imprinted X inactivation during early mouse developmentScience2004303565864464914671313
- ChengMKDistecheCMSilence of the fathers: early X inactivationBioessays200426882182415273983
- LucchesiJCKellyWGPanningBChromatin remodeling in dosage compensationAnn Rev Genet20053961565116285873
- Zandman-GoddardGPeevaEShoenfeldYGender and autoimmunityAutoimmun Rev20076636637217537382
- McCombePAGreerJMMackayIRSexual dimorphism in autoimmune diseaseCurr Mol Med2009991058107919747114
- GreerJMMcCombePARole of gender in multiple sclerosis: clinical effects and potential molecular mechanismsJ Neuroimmunol20112341–271821474189
- IslamTGaudermanWJCozenWHamiltonASBurnettMEMackTMDifferential twin concordance for multiple sclerosis by latitude of birthplaceAnn Neurol2006601566416685699
- RistoriGCannoniSStaziMAMultiple sclerosis in twins from continental Italy and Sardinia: a nationwide studyAnn Neurol2006591273416240370
- RedondoMJJeffreyJFainPREisenbarthGSOrbanTConcordance for islet autoimmunity among monozygotic twinsN Engl J Med2008359262849285019109586
- JacobsKBYeagerMZhouWDetectable clonal mosaicism and its relationship to aging and cancerNat Genet201244665165822561519
- LaurieCCLaurieCARiceKDetectable clonal mosaicism from birth to old age and its relationship to cancerNat Genet201244664265022561516
- JavierreBMFernandezAFRichterJChanges in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosusGenome Res201020217017920028698
- BaranziniSEMudgeJvan VelkinburghJCGenome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosisNature201046472931351135620428171
- BellCGTeschendorffAERakyanVKMaxwellAPBeckSSavageDAGenome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitusBMC Med Genomics201033320687937
- BreitlingLPYangRKornBBurwinkelBBrennerHTobacco-smoking-related differential DNA methylation: 27K discovery and replicationAm J Hum Genet201188445045721457905
- RakyanVKDownTABaldingDJBeckSEpigenome-wide association studies for common human diseasesNat Rev Genet201112852954121747404
- GervinKVigelandMDMattingsdalMDNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: identification of epigenetically dysregulated genesPLoS Genet201281e100245422291603
- RakyanVKBeyanHDownTAIdentification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosisPLoS Genet201179e100230021980303
- HeijmansBTMillJCommentary: The seven plagues of epigenetic epidemiologyInt J Epidemiol2012411747822269254
- WilsonCBRowellESekimataMEpigenetic control of T-helper-cell differentiationNat Rev Immunol2009929110519151746
- AuneTMCollinsPLChangSEpigenetics and T helper 1 differentiationImmunology2009126329930519178593
- BergmanYCedarHEpigenetic control of recombination in the immune systemSemin Immunol201022632332920832333
- CedarHBergmanYEpigenetics of haematopoietic cell developmentNat Rev Immunol201111747848821660052
- Fernández-MoreraJLCalvaneseVRodríguez-RoderoSMenéndez-TorreEFragaMFEpigenetic regulation of the immune system in health and diseaseTissue Antigens201076643143921058938
- JansonPCWinerdalMEWinqvistOAt the crossroads of T helper lineage commitment-Epigenetics points the wayBiochim Biophys Acta20091790990691919162128
- LalGBrombergJSEpigenetic mechanisms of regulation of Foxp3 expressionBlood2009114183727373519641188
- LeeGRKimSTSpilianakisCGFieldsPEFlavellRAT helper cell differentiation: regulation by cis elements and epigeneticsImmunity200624436937916618596
- PangKCDingerMEMercerTRGenome-wide identification of long noncoding RNAs in CD8+ T cellsJ Immunol2009182127738774819494298
- SawalhaAHEpigenetics and T-cell immunityAutoimmunity200841424525218432405
- Vanden BergheWNdlovuMNHoya-AriasRDijsselbloemNGerloSHaegemanGKeeping up NF-kappaB appearances: epigenetic control of immunity or inflammation-triggered epigeneticsBiochem Pharmacol20067291114113116934762
- WilsonCBMakarKWShnyrevaMFitzpatrickDRDNA methylation and the expanding epigenetics of T cell lineage commitmentSemin Immunol200517210511915737572
- AllanRSZuevaECammasFAn epigenetic silencing pathway controlling T helper 2 cell lineage commitmentNature2012 Epub July 4.
- BlümlSZupkovitzGKirchbergerSEpigenetic regulation of dendritic cell differentiation and function by oxidized phospholipidsBlood2009114275481548919864645
- SubrahmanyamRSenREpigenetic features that regulate IgH locus recombination and expressionCurr Top Microbiol Immunol2012356396321779986
- KohASKingstonREBenoistCMathisDGlobal relevance of Aire binding to hypomethylated lysine-4 of histone-3Proc Natl Acad Sci U S A201010729130161302120615959
- WilliamsGTMourtada-MaarabouniMFarzanehFA critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytesBiochem Soc Trans201139248248621428924
- KinoTHurtDEIchijoTNaderNChrousosGPNoncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptorSci Signal20103107ra820124551
- AshwellJDLuFWVacchioMSGlucocorticoids in T cell development and functionAnnu Rev Immunol20001830934510837061
- LiQJChauJEbertPJmiR-181a is an intrinsic modulator of T cell sensitivity and selectionCell2007129114716117382377
- BelkayaSSilgeRLHooverARDynamic modulation of thymic microRNAs in response to stressPloS One2011611e2758022110677
- LiBSamantaASongXFOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repressionProc Natl Acad Sci U S A2007104114571457617360565
- TaoRde ZoetenEFÖzkaynakEDeacetylase inhibition promotes the generation and function of regulatory T cellsNat Med200713111299130717922010
- BettiniMLPanFBettiniMLoss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiencyImmunity201236571773022579476
- BeyerMThabetYMüllerRURepression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiationNat Immunol201112989890721841785
- ZhouLParkJJZhengQDongZMiQMicroRNAs are key regulators controlling iNKT and regulatory T-cell development and functionCell Mol Immunol20118538038721822298
- SongWTaiYTTianZHDAC inhibition by LBH589 affects the phenotype and function of human myeloid dendritic cellsLeukemia201125116116821102427
- IliopoulosDKavousanakiMIoannouMBoumpasDVerginisPThe negative costimulatory molecule PD-1 modulates the balance between immunity and tolerance via miR-21Eur J Immunol20114161754176321469086
- GialitakisMKretsovaliASpilianakisCCoordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin ANucleic Acids Res200634376577216452299
- RamagopalanSVEbersGCMultiple sclerosis: major histocompatibility complexity and antigen presentationGenome Med200911110519895714
- ChaoMJRamagopalanSVHerreraBMEpigenetics in multiple sclerosis susceptibility: difference in transgenerational risk localizes to the major histocompatibility complexHum Mol Genet200918226126619098025
- ChaoMJRamagopalanSVHerreraBMMHC transmission: insights into gender bias in MS susceptibilityNeurology201176324224621209377
- HedrichCMTsokosGCEpigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseasesTrends Mol Med2011171271472421885342
- RenaudineauYGaraudSLe DantecCAlonso-RamirezRDaridonCYouinouPAutoreactive B cells and epigeneticsClin Rev Allergy Immunol2010391859419644775
- ListonAAutoimmunity: beyond the immune systemImmunol Cell Biol200886429529618301386
- LonyaiAKodamaSBurgerDFaustmanDLFetal Hox11 expression patterns predict defective target organs: a novel link between developmental biology and autoimmunityImmunol Cell Biol200886430130918301381
- KarouzakisEGayREMichelBAGaySNeidhartMDNA hypomethylation in rheumatoid arthritis synovial fibroblastsArthritis Rheum200960123613362219950268
- HuberLCBrockMHemmatazadHHistone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patientsArthritis Rheum20075641087109317393417
- MastronardiFGNoorAWoodDDPatonTMoscarelloMAPeptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylatedJ Neurosci Res20078592006201617469138
- CiavattaDJYangJPrestonGAEpigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitisJ Clin Invest201012093209321920714105
- WenZKXuWXuLDNA hypomethylation is crucial for apoptotic DNA to induce systemic lupus erythematosus-like autoimmune disease in SLE-non-susceptible miceRheumatology200746121796180318032537
- DiekerJMullerSEpigenetic Histone Code and AutoimmunityClin Rev Allergy Immunol2010391788419662539
- PlauéSMullerSvan RegenmortelMHA branched, synthetic octapeptide of ubiquitinated histone H2A as target of autoantibodiesJ Exp Med19891695160716172541220
- DiekerJWFransenJHvan BavelCCApoptosis-induced acetylation of histones is pathogenic in systemic lupus erythematosusArthritis Rheum20075661921193317530637
- KlipHVerloopJvan GoolJDKosterMEBurgerCWvan LeeuwenFEOMEGA Project GroupHypospadias in sons of women exposed to diethylstilbestrol in utero: a cohort studyLancet200235993121102110711943257
- AlonsoAHernánMATemporal trends in the incidence of multiple sclerosis: a systematic reviewNeurology200871212913518606967
- HirstMMarraMAEpigenetics and human diseaseInt J Biochem Cell Biol200941113614618852064
- OrtonSMHerreraBMYeeIMCanadian Collaborative Study GroupSex ratio of multiple sclerosis in Canada: a longitudinal studyLancet Neurol200651193293617052660
- OsoegawaMKiraJFukazawaTResearch Committee of Neuroimmunological DiseasesTemporal changes and geographical differences in multiple sclerosis phenotypes in Japanese: nationwide survey results over 30 yearsMult Scler200915215917318987106
- FurszyferJKurlandLTMcConaheyWMWoolnerLBElvebackLREpidemiologic aspects of Hashimoto’s thyroiditis and Graves’ disease in Rochester, Minnesota (1935–1967), with special reference to temporal trendsMetabolism19722131972045066850
- PoirierLAThe effects of diet, genetics and chemicals on toxicity and aberrant DNA methylation: an introductionJ Nutr2002132Suppl 82336S2339S12163688
- McGowanPOMeaneyMJSzyfMDiet and the epigenetic (re) programming of phenotypic differences in behaviorBrain Res20081237122418694740
- HeijmansBTTobiEWSteinADPersistent epigenetic differences associated with prenatal exposure to famine in humansProc Natl Acad Sci U S A200810544170461704918955703
- van EngelandMWeijenbergMPRoemenGMEffects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancerCancer Res200363123133313712810640
- de VogelSBongaertsBWWoutersKAAssociations of dietary methyl donor intake with MLH1 promoter hypermethylation and related molecular phenotypes in sporadic colorectal cancerCarcinogenesis20082991765177318339680
- KuneGWatsonLColorectal cancer protective effects and the dietary micronutrients folate, methionine, vitamins B6, B12, C, E, selenium, and lycopeneNutr Cancer2006561112117176213
- MasSLafuenteMJCrescentiALower specific micronutrient intake in colorectal cancer patients with tumors presenting promoter hypermethylation in p16(INK4a), p4(ARF) and hMLH1Anticancer Res20072721151115617465256
- HardyTMTollefsbolTOEpigenetic diet: impact on the epigenome and cancerEpigenomics20113450351822022340
- SelmiCTsuneyamaKNutrition, geoepidemiology, and autoimmunityAutoimmun Rev201095A267A27019969106
- ProcacciniCCarboneFGalganiMObesity and susceptibility to autoimmune diseasesExpert Rev Clin Immunol20117328729421595595
- GardenerHMungerKLChitnisTMichelsKBSpiegelmanDAscherioAPrenatal and perinatal factors and risk of multiple sclerosisEpidemiology200920461161819333127
- MungerKLChitnisTAscherioABody size and risk of MS in two cohorts of US womenNeurology200973191543155019901245
- Weinstock-GuttmanBZivadinovRMahfoozNSerum lipid profiles are associated with disability and MRI outcomes in multiple sclerosisJ Neuroinflammation2011812721970791
- MatareseGCarrieriPBLa CavaALeptin increase in multiple sclerosis associates with reduced number of CD4+CD25+ regulatory T cellsProc Natl Acad Sci U S A2005102145150515515788534
- CampiónJMilagroFIMartínezJAIndividuality and epigenetics in obesityObes Rev200910438339219413700
- McLeodJGHammondSRHallpikeJFEpidemiology of multiple sclerosis in Australia. With NSW and SA survey resultsMed J Aust199416031171228295576
- StaplesJAPonsonbyALLimLLMcMichaelAJEcologic analysis of some immune-related disorders, including type 1 diabetes, in Australia: latitude, regional ultraviolet radiation, and disease prevalenceEnviron Health Perspect2003111451852312676609
- AlamanosYVoulgariPVDrososAAIncidence and prevalence of rheumatoid arthritis, based on the 1987 American College of Rheumatology criteria: a systematic reviewSemin Arthritis Rheum200636318218817045630
- HengstmanGJvan VenrooijWJVencovskyJMoutsopoulosHMvan EngelenBGThe relative prevalence of dermatomyositis and polymyositis in Europe exhibits a latitudinal gradientAnn Rheum Dis200059214114210666171
- BeretichBDBeretichTMExplaining multiple sclerosis prevalence by ultraviolet exposure: a geospatial analysisMult Scler200915889189819667017
- BecklundBRSeversonKSVangSVDeLucaHFUV radiation suppresses experimental autoimmune encephalomyelitis independent of vitamin D productionProc Natl Acad Sci U S A2010107146418642320308557
- DongZBodeAMThe role of histone H3 phosphorylation (Ser10 and Ser28) in cell growth and cell transformationMol Carcinog200645641642116637065
- StaibanoSMascoloMSianoMIlardiGDe RosaGGenetic, epigenetic and molecular changes in melanoma: a new paradigm for biological classificationMurphMResearch on Melanoma: A Glimpse into Current Directions and Future TrendsRijekaInTech20113568 Available from: http://www.intechopen.com/books/research-on-melanoma-a-glimpse-into-current-directions-and-future-trends/genetic-epigenetic-and-molecular-changes-in-melanoma-a-new-paradigm-for-biological-classification. Accessed July 13, 2012.
- HallHIMayDSLewRAKohHKNadelMSun protection behaviors of the US white populationPrev Med19972644014079245656
- Thomas-AhnerJMWulffBCToberKLKusewittDFRiggenbachJAOberyszynTMGender differences in UVB-induced skin carcinogenesis, inflammation, and DNA damageCancer Res20076773468347417389759
- KarlicHVargaFImpact of vitamin D metabolism on clinical epigeneticsClin Epigenetics201121556122704269
- SongYFleetJC1,25 dihydroxycholecalciferol-mediated calcium absorption and gene expression are higher in female than in male miceJ Nutr200413481857186115284366
- KragtJvan AmerongenBKillesteinJHigher levels of 25-hydroxyvitamin D are associated with a lower incidence of multiple sclerosis only in womenMult Scler200915191518701572
- BigazziPEAutoimmunity and heavy metalsLupus1994364494537704000
- HemdanNYAEmmrichFFaberSLehmannJSackUAlterations of Th1/Th2 reactivity by heavy metals possible consequences include induction of autoimmune diseasesAnn N Y Acad Sci2007110912913717785298
- HouLZhangXWangDBaccarelliAEnvironmental chemical exposures and human epigeneticsInt J Epidemiol20114117910522253299
- Martinez-ZamudioRHaHCEnvironmental epigenetics in metal exposureEpigenetics20116782082721610324
- ArnsonYShoenfeldYAmitalHEffects of tobacco smoke on immunity, inflammation and autoimmunityJ Autoimmun2010343J258J26520042314
- SuterMMaJHarrisASMaternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expressionEpigenetics20116111284129421937876
- YungRLRichardsonBCDrug-induced lupusRheum Dis Clin North Am199420161867512273
- LeeBHYegnasubramanianSLinXNelsonWGProcainamide is a specific inhibitor of DNA methyltransferase 1J Biol Chem200528049407494075616230360
- GorelikGFangJYWuASawalhaAHRichardsonBImpaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupusJ Immunol200717985553556317911642
- RiegerRLeungPSJeddelohMRIdentification of 2-nonynoic acid, a cosmetic component, as a potential trigger of primary biliary cirrhosisJ Autoimmun200627171616876981
- PrinsGSEstrogen imprinting: when your epigenetic memories come back to haunt youEndocrinology2008149125919592119022898
- Guerrero-BosagnaCSabatPValladaresLEnvironmental signaling and evolutionary change: can exposure of pregnant mammals to environmental estrogens lead to epigenetically induced evolutionary changes in embryos?Evol Dev20057434135015982371
- DolinoyDCWeidmanJRWaterlandRAJirtleRLMaternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenomeEnviron Health Perspect2006114456757216581547
- StrohsnitterWCNollerKLTroisiRAutoimmune disease incidence among women prenatally exposed to DiethylstilbestrolJ Rheumatol201037102167217320634240
- CampesiISannaMZinelluAOral contraceptives modify DNA methylation and monocyte-derived macrophage functionBiol Sex Differ20123422284681
- HellwigKSchimrigkSBesteCMullerTGoldRIncrease in relapse rate during assisted reproduction technique in patients with multiple sclerosisEur Neurol2009612656819039223
- KatariSTuranNBibikovaMDNA methylation and gene expression differences in children conceived in vitro or in vivoHum Mol Genet200918203769377819605411
- van MontfoortAPHanssenLLde SutterPVivilleSGeraedtsJPde BoerPAssisted reproduction treatment and epigenetic inheritanceHum Reprod Update201218217119722267841
- KriegAMGourleyMFPerlAEndogenous retroviruses: potential etiologic agents in autoimmunityFASEB J199268253725441592206
- NakkuntodJAvihingsanonYMutiranguraAHirankarnNHypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patientsClin Chim Acta201141215–161457146121496453
- IssaJPOttavianoYLCelanoPHamiltonSRDavidsonNEBaylinSBMethylation of the oestrogen receptor CpG island links ageing and neoplasia in human colonNat Genet1994745365407951326
- SargBKoutzamaniEHelligerWRundquistILindnerHHPostsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with agingJ Biol Chem200227742391953920112154089
- FragaMFBallestarEPazMFEpigenetic differences arise during the lifetime of monozygotic twinsProc Natl Acad Sci U S A200510230106041060916009939
- BellJTTsaiPCYangTPEpigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing populationPLoS Genet201284e100262922532803
- RakyanVKDownTAMaslauSHuman aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domainsGenome Res201020443443920219945
- BernsteinBEMikkelsenTSXieXA bivalent chromatin structure marks key developmental genes in embryonic stem cellsCell2006125231532616630819
- TeschendorffAEMenonUGentry-MaharajAAge-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancerGenome Res201020444044620219944
- JavierreBMHernandoHBallestarEEnvironmental triggers and epigenetic deregulation in autoimmune diseaseDiscov Med2011126753554522204770
- EssexMJThomas BoyceWHertzmanCEpigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescenceChild Dev2011 Epub Sep 2.
- WeaverICCervoniNChampagneFAEpigenetic programming by maternal behaviorNat Neurosci20047884785415220929
- McCarthyMMAugerAPBaleTLThe epigenetics of sex differences in the brainJ Neurosci20092941128151282319828794
- BestJDCareyNThe epigenetics of normal pregnancyObstet Med2011 Epub Dec 15.
- D’hoogheMBNagelsGUitdehaagBMLong-term effects of childbirth in MSJ Neurol Neurosurg Psychiatry2010811384119939856
- RunmarkerBAndersenOPregnancy is associated with a lower risk of onset and a better prognosis in multiple sclerosisBrain1995118Pt 12532617895009
- VerdruPTheysPD’HoogheMBCartonHPregnancy and multiple sclerosis: the influence on long term disabilityClinical Neurol Neurosurg19949613841
- GleesonMBishopNCStenselDJLindleyMRMastanaSSNimmoMAThe anti-inflammatory effects of exercise: mechanisms and implications for the prevention and treatment of diseaseNat Rev Immunol201111960761521818123
- DalgasUStenagerEExercise and disease progression in multiple sclerosis: can exercise slow down the progression of multiple sclerosis?Ther Adv Neurol Disord201252819522435073
- BarrèsRYanJEganBAcute exercise remodels promoter methylation in human skeletal muscleCell Metab201215340541122405075
- McGeeSLFairlieEGarnhamAPHargreavesMExercise-induced histone modifications in human skeletal muscleJ Physiol2009587Pt 245951595819884317
- BallestarEEstellerMRichardsonBCThe epigenetic face of systemic lupus erythematosusJ Immunol2006176127143714716751355
- LuQWuATesmerLRayDYousifNRichardsonBDemethylation of CD40 LG on the inactive X in T cells from women with lupusJ Immunol200717996352635817947713
- AlvesNLArosaFAvan LierRACommon gamma chain cytokines: dissidence in the detailsImmunol Lett2007108211312017194484
- BennettCLChristieJRamsdellFThe immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3Nat Genet2001271202111137993
- SpurrNKGoodfellowPNDochertyAJChromosomal assignment of the gene encoding the human tissue inhibitor of metalloproteinases to Xp11.1-p11.4Ann Hum Genet198751Pt 31891943688834
- ZehntnerSPBourbonnièreLMooreCSX-linked inhibitor of apoptosis regulates T cell effector functionJ Immunol2007179117553756018025200
- Møller-LarsenSNyegaardMHaagerupAVestboJKruseTABørglumADAssociation analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disordersThorax200863121064106918682521
- ChitnisSMonteiroJGlassDThe role of X-chromosome inactivation in female predisposition to autoimmunityArthritis Res20002539940611056674
- BrixTHKnudsenGPKristiansenMKyvikKOOrstavikKHHegedüsLHigh frequency of skewed X-chromosome inactivation in females with autoimmune thyroid disease: a possible explanation for the female predisposition to thyroid autoimmunityJ Clin Endocrinol Metab.200590115949595316105963
- OzcelikTUzEAkyerliCBEvidence from autoimmune thyroiditis of skewed X-chromosome inactivation in female predisposition to autoimmunityEur J Hum Genet200614679179716596118
- OzbalkanZBagişlarSKirazSSkewed X chromosome inactivation in blood cells of women with sclerodermaArthritis Rheum20055251564157015880831
- HuangQParfittAGrennanDMManoliosNX-chromosome inactivation in monozygotic twins with systemic lupus erythematosusAutoimmunity199726285939546817
- KnudsenGPGender bias in autoimmune diseases: X chromosome inactivation in women with multiple sclerosisJ Neurol Sci20092861–2434619411081
- BoyleAPDavisSShulhaHPHigh-resolution mapping and characterization of open chromatin across the genomeCell2008132231132218243105
- HesselberthJRChenXZhangZGlobal mapping of protein-DNA interactions in vivo by digital genomic footprintingNat Methods20096428328919305407
- ParkPJChIP-seq: advantages and challenges of a maturing technologyNat Rev Genet2009101066968019736561
- LairdPWPrinciples and challenges of genomewide DNA methylation analysisNat Rev Genet201011319120320125086
- BirneyEChromatin and heritability: how epigenetic studies can complement genetic approachesTrends Genet201127517217621414682
- BarskiACuddapahSCuiKHigh-resolution profiling of histone methylations in the human genomeCell2007129482383717512414
- CamposEIReinbergDHistones: annotating chromatinAnn Rev Genet20094355959919886812
- KochCMAndrewsRMFlicekPThe landscape of histone modifications across 1% of the human genome in five human cell linesGenome Res200717669170717567990
- RosenfeldJAWangZSchonesDEZhaoKDeSalleRZhangMQDetermination of enriched histone modifications in non-genic portions of the human genomeBMC Genomics20091014319335899
- ZhouVWGorenABernsteinBECharting histone modifications and the functional organization of mammalian genomesNat Rev Genet201112171821116306
- LennartssonAEkwallKHistone modification patterns and epigenetic codesBiochim Biophys Acta20091790986386819168116
- HuffJTPlocikAMGuthrieCYamamotoKRReciprocal intronic and exonic histone modification regions in humansNat Struct Mol Biol201017121495149921057525
- HirstCIngramGPickersgillTSwinglerRCompstonDARobertsonNPIncreasing prevalence and incidence of multiple sclerosis in South East WalesJ Neurol Neurosurg Psychiatry200980438639118931003
- WeinshenkerBGBassBRiceGPThe natural history of multiple sclerosis: a geographically based study. I. Clinical course and disabilityBrain1989112Pt 11331462917275
- EbersGCBulmanDESadovnickADA population-based study of multiple sclerosis in twinsN Engl J Med198631526163816423785335
- WillerCJDymentDARischNJSadovnickADEbersGCThe Canadian Collaborative Study GroupTwin concordance and sibling recurrence rates in multiple sclerosisProc Natl Acad Sci U S A200310022128771288214569025
- HansenTSkyttheAStenagerEPetersenHCBrønnum-HansenHKyvikKOConcordance for multiple sclerosis in Danish twins: an update of a nationwide studyMult Scler200511550451016193885
- KuusistoHKaprioJKinnunenELuukkaalaTKoskenvuoMElovaaraIConcordance and heritability of multiple sclerosis in Finland: study on a nationwide series of twinsEur J Neurol200815101106111018727671
- OstmanJLonnbergGArnqvistHJGender differences and temporal variation in the incidence of type 1 diabetes: results of 8012 cases in the nationwide Diabetes Incidence Study in Sweden 1983–2002J Intern Med2008263438639418205768
- EatonWWRoseNRKalaydjianAPedersenMGMortensenPBEpidemiology of autoimmune diseases in DenmarkJ Autoimmun20072911917582741
- KaprioJTuomilehtoJKoskenvuoMConcordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in FinlandDiabetologia19923511106010671473616
- MetcalfeKAHitmanGARoweREConcordance for type 1 diabetes in identical twins is affected by insulin genotypeDiabetes Care200124583884211347740
- KyvikKOGreenABeck-NielsenHConcordance rates of insulin dependent diabetes mellitus: a population based study of young Danish twinsBMJ199531170109139177580548
- McCartyDJManziSMedsgerTAJrRamsey-GoldmanRLaPorteREKwohCKIncidence of systemic lupus erythematosus. Race and gender differencesArthritis Rheum1995389126012707575721
- DeapenDEscalanteAWeinribLA revised estimate of twin concordance in systemic lupus erythematosusArthritis Rheum19923533113181536669
- JärvinenPKaprioJMäkitaloRKoskenvuoMAhoKSystemic lupus erythematosus and related systemic diseases in a nationwide twin cohort: an increased prevalence of disease in MZ twins and concordance of disease featuresJ Intern Med1992231167721732401
- GrennanDMParfittAManoliosNFamily and twin studies in systemic lupus erythematosusDis Markers199713293989160184
- GonzalezAMaradit KremersHCrowsonCSThe widening mortality gap between rheumatoid arthritis patients and the general populationArthritis Rheum200756113583358717968923
- WinchesterRJGenetic aspects of rheumatoid arthritisSpringer Semin Immunopathol198142891026459655
- AhoKKoskenvuoMTuominenJKaprioJOccurrence of rheumatoid arthritis in a nationwide series of twinsJ Rheumatol19861358999023820198
- SilmanAJMacGregorAJThomsonWTwin concordance rates for rheumatoid arthritis: results from a nationwide studyBr J Rheumatol199332109039078402000
- KrassasGETziomalosKPontikidesNLewyHLaronZSeasonality of month of birth of patients with Graves’ and Hashimoto’s diseases differ from that in the general populationEur J Endocrinol2007156663163617535862
- BrixTHKyvikKOChristensenKHegedüsLEvidence for a major role of heredity in Graves’ disease: a population-based study of two Danish twin cohortsJ Clin Endocrinol Metab200186293093411158069
- RingoldDANicoloffJTKeslerMDavisHHamiltonAMackTFurther evidence for a strong genetic influence on the development of autoimmune thyroid disease: the California twin studyThyroid200212864765312225632
- KaplanMMGershwinMEPrimary biliary cirrhosisN Engl J Med2005353121261127316177252
- SelmiCMayoMJBachNPrimary biliary cirrhosis in monozygotic and dizygotic twins: genetics, epigenetics, and environmentGastroenterology2004127248549215300581
- NaldiLChatenoudLBelloniAMedical history, drug exposure and the risk of psoriasis. Evidence from an Italian case-control studyDermatology2008216212513018216474
- DuffyDLSpelmanLSMartinNGPsoriasis in Australian twinsJ Am Acad Dermatol19932934284348349859
- KruegerGGDuvicMEpidemiology of psoriasis: clinical issuesJ Invest Dermatol1994102614S18S8006427
- MantegazzaRBaggiFAntozziCMyasthenia gravis (MG): epidemiological data and prognostic factorsAnn N Y Acad Sci200399841342314592909
- RamanujamRPirskanenRRamanujamSHammarströmLUtilizing twins concordance rates to infer the predisposition to myasthenia gravisTwin Res Hum Genet201114212913621425894
- WillREdmundsLElswoodJCalinAIs there sexual inequality in ankylosing spondylitis? A study of 498 women and 1202 menJ Rheumatol19901712164916522084239
- PedersenOBSvendsenAJEjstrupLSkyttheAHarrisJRJunkerPAnkylosing spondylitis in Danish and Norwegian twins: occurrence and the relative importance of genetic vs environmental effectors in disease causationScand J Rheumatol200837212012618415769
- BaladaEOrdi-RosJVilardell-TarrésMDNA methylation and systemic lupus erythematosusAnn N Y Acad Sci2007110812713617893979
- RenaudineauYYouinouPEpigenetics and autoimmunity, with special emphasis on methylationKeio J Med2011601101621460598
- ZoualiMEpigenetics in lupusAnn N Y Acad Sci2011121715416521251010
- GaraudSLe DantecCJousse-JoulinSIL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylationJ Immunol200918295623563219380809
- PauleyKMSatohMChanALBubbMRReevesWHChanEKUpregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patientsArthritis Res Ther2008104R10118759964
- HuNQiuXLuoYAbnormal histone modification patterns in lupus CD4+ T cellsJ Rheumatol200835580481018398941
- LuoYLiYSuYAbnormal DNA methylation in T cells from patients with subacute cutaneous lupus erythematosusBr J Dermatol2008159482783318644019
- LeiWLuoYLeiWAbnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositisScand J Rheumatol200938536937419444718
- SunahoriKJuangYTKyttarisVCTsokosGCPromoter hypomethylation results in increased expression of protein phosphatase 2A in T cells from patients with systemic lupus erythematosusJ Immunol201118674508451721346232
- JanuchowskiRWudarskiMChwalińska-SadowskaHJagodzinskiPPPrevalence of ZAP-70, LAT, SLP-76, and DNA methyltransferase 1 expression in CD4+ T cells of patients with systemic lupus erythematosusClin Rheumatol2008271212717492476
- YinHZhaoMWuXHypomethylation and overexpression of CD70 (TNFSF7) in CD4+ T cells of patients with primary Sjögren’s syndromeJ Dermatol Sci201059319820320724115
- JiangHXiaoRLianXDemethylation of TNFSF7 contributes to CD70 overexpression in CD4+ T cells from patients with systemic sclerosisClin Immunol20121431394422306512
- LiuYChenYRichardsonBDecreased DNA methyltransferase levels contribute to abnormal gene expression in “senescent” CD4(+) CD28(−) T cellsClin Immunol2009132225726519394279
- FulciVScappucciGSebastianiGDmiR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritisHum Immunol201071220621119931339
- JansonPCLintonLBBergmanEAProfiling of CD4+ T Cells with epigenetic immune lineage analysisJ Immunol201118619210221131423
- JunkerAPathophysiology of translational regulation by microRNAs in multiple sclerosisFEBS Letters2011585233738374621453702
- LleoALiaoJInvernizziPImmunoglobulin M levels inversely correlate with CD40 ligand promoter methylation in patients with primary biliary cirrhosisHepatology201255115316021898485
- LuQWuARayDDNA methylation and chromatin structure regulate T cell perforin gene expressionJ Immunol2003170105124513212734359
- DaiYHuangYSTangMMicroarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patientsLupus2007161293994618042587
- MiaoFSmithDDZhangLMinAFengWNatarajanRLymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetesDiabetes200857123189319818776137
- van BavelCCDiekerJMullerSApoptosis-associated acetylation on histone H2B is an epitope for lupus autoantibodiesMol Immunol2009472–351151619747733
- NakasaTMiyakiSOkuboAExpression of microRNA-146 in rheumatoid arthritis synovial tissueArthritis Rheum20085851284129218438844
- StanczykJOspeltCKarouzakisEAltered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activationArthritis Rheum201163237338121279994
- TakamiNOsawaKMiuraYHypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cellsArthritis Rheum200654377978716508942
- Sánchez-PernauteOOspeltCNeidhartMGaySEpigenetic clues to rheumatoid arthritisJ Autoimmun2008301–2122018155418
- PadgettKALanRYLeungPCPrimary biliary cirrhosis is associated with altered hepatic microRNA expressionJ Autoimmun2009323–424625319345069
- AlevizosIIlleiGGMicroRNAs in Sjögren’s syndrome as a prototypic autoimmune diseaseAutoimmun Rev20109961862120457282
- JoyceCEZhouXXiaJDeep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAomeHum Mol Genet201120204025404021807764
- RobersonEDLiuYRyanCA subset of methylated CpG sites differentiate psoriatic from normal skinJ Invest Dermatol20121323 Pt 158359222071477
- RuchusatsawatKWongpiyabovornJShuangshotiSHirankarnNMutiranguraASHP-1 promoter 2 methylation in normal epithelial tissues and demethylation in psoriasisJ Mol Med (Berl)200684217518216389548
- HuynhJLCasacciaPDefining the chromatin landscape in demyelinating disordersNeurobiol Dis2010391475219853663
- KuboMCzuwara-LadykowskaJMoussaOPersistent downregulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skinAm J Pathol2003163257158112875977
- LugerKMäderAWRichmondRKSargentDFRichmondTJCrystal structure of the nucleosome core particle at 2.8 A resolutionNature199738966482512609305837