Abstract
Background
Dengue illness is one of the important mosquito-borne viral diseases in tropical and subtropical regions. Four serotypes of dengue virus (DENV-1, DENV-2, DENV-3, and DENV-4) are classified in the Flavivirus genus of the family Flaviviridae. We prepared monoclonal antibodies against DENV capsid protein from mice immunized with DENV-2 and determined the cross-reactivity with each serotype of DENV and Japanese encephalitis virus.
Methods and results
To clarify the relationship between the cross-reactivity of monoclonal antibodies and the diversity of these viruses, we examined the situations of flaviviruses by analyses of phylogenetic trees. Among a total of 60 prepared monoclonal antibodies specific for DENV, five monoclonal antibodies stained the nuclei of infected cells and were found to be specific to the capsid protein. Three were specific to DENV-2, while the other two were cross-reactive with DENV-2 and DENV-4. No monoclonal antibodies were cross-reactive with all four serotypes. Phylogenetic analysis of DENV amino acid sequences of the capsid protein revealed that DENV-2 and DENV-4 were clustered in the same branch, while DENV-1 and DENV-3 were clustered in the other branch. However, these classifications of the capsid protein were different from those of the envelope and nonstructural 1 proteins. Phylogenetic distances between the four serotypes of DENV were as different as those of other flaviviruses, such as Japanese encephalitis virus and West Nile virus. Large variations in the DENV serotypes were comparable with the differences between species of flavivirus. Furthermore, the diversity of flavivirus capsid protein was much greater than that of envelope and nonstructural 1 proteins.
Conclusion
In this study, we produced specific monoclonal antibodies that can be used to detect DENV-2 capsid protein, but not a cross-reactive one with all serotypes of DENV capsid protein. The high diversity of the DENV capsid protein sequence by phylogenetic analysis supported the low cross-reactivity of monoclonal antibodies against DENV capsid protein.
Introduction
Dengue illness, such as dengue fever and dengue hemorrhagic fever/dengue shock syndrome, is one of the important mosquito-borne viral diseases in tropical and subtropical regions.Citation1 Four serotypes of dengue virus (DENV-1 to DENV-4) are classified in the genus Flavivirus of the family Flaviviridae. This also includes Japanese encephalitis virus, West Nile virus, and yellow fever virus.Citation2
The 11 kb DENV genome is translated into a single polyprotein, which is subsequently processed by proteases into structural and nonstructural proteins. Three structural proteins, i.e. capsid, premembrane/membrane, and envelope, make up the virus particle.Citation3 The DENV capsid protein is a relatively small, highly positively charged 12 kDa protein and an essential factor during virion assembly.Citation4,Citation5 Interestingly, DENV capsid protein is found in the nucleus and nucleoli of infected cells as early as 6 hours after infection, well before formation of infectious virus.Citation6,Citation7 Previous reports have suggested nucleolar accumulation of DENV capsid protein may result from its interaction with RNA or nucleoli proteins related to regulation of ribosome synthesis, mRNA processing, and DNA replication.Citation5,Citation8,Citation9 This nuclear localization of capsid protein appears to be conserved among flaviviruses, including West Nile virus, Japanese encephalitis virus, Kunjin virus, and hepatitis C virus.Citation10–Citation13
In general, antibodies against the capsid proteins of viral particles, such as human immunodeficiency virus (HIV) Gag protein and influenza virus nucleoprotein, have been used in antigen-capture diagnosis kitsCitation14,Citation15 because of their wide range of cross-reactivity, even with other subtypes, compared with that of the envelope proteins.Citation16,Citation17 Similar approaches using antigen-capture diagnostic or enzyme-linked immunoassay kits have been used for other viruses, such as Rift Valley fever virus,Citation18 hepatitis B virus,Citation19 hepatitis C virus,Citation20 and Ebola virus.Citation21 Such viral structural capsid proteins are located inside, not on the surface, of viral particles and the amount of internal protein exists at a higher concentration than that of surface protein. Therefore, use of monoclonal antibodies against the internal structural protein could have advantages for developing rapid diagnostic kits. In addition, expression of DENV capsid protein has been detected at a very early stage in infected mammalian cells.Citation33 Thus, if antibodies against DENV capsid protein were also widely cross-reactive among all four serotypes of DENV, like those against HIV Gag protein and influenza virus nucleoprotein, monoclonal antibodies recognizing common antigenic regions on DENV capsid proteins for all four DENV serotypes could be highly useful for developing a rapid diagnostic test kit widely cross-reactive with all four serotypes in the acute phase. However, monoclonal antibodies to the capsid protein has not been used for development of a diagnostic kit to detect this viral infection, and monoclonal antibodies to DENV nonstructural 1 protein have been used instead for developing diagnostic kits.Citation22,Citation23
In this study, we attempted to produce monoclonal antibodies against DENV capsid protein that could be cross-reactive with all four serotypes of DENV. By immunizing BALB/c mice with DENV-2, we obtained a total of five hybridoma clones producing specific antibodies against DENV capsid protein. We hypothesized that a monoclonal antibody against DENV capsid protein would recognize common antigenic regions on the capsid proteins of all four DENV serotypes and be very useful for developing a diagnostic test kit widely cross-reactive with all four serotypes. The purpose of this study was to clarify the relationship between the cross-reactivity of monoclonal antibodies and the diversity of these viruses, and we examined the situation of flaviviruses by analysis of phylogenetic trees.
Materials and methods
Cells and viruses
Vero cells (from an African green monkey kidney epithelial cell line) and B-7 cells (from a BALB/c mouse cell line)Citation24 were maintained in Eagle’s Minimum Essential Medium (MEM, HyClone Laboratories Inc, Logan, UT) supplemented with 10% fetal calf serum (HyClone Laboratories Inc). PAI cells (from a mouse myeloma cell line) were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640, HyClone Laboratories Inc) medium supplemented with 10% fetal calf serum. All cell lines were cultured at 37°C in a 5% CO2 incubator. C6/36 cells (from an Aedes albopictus cell line) were grown in Leibovitz-15 medium (HyClone Laboratories, Inc) supplemented with 0.3% tryptose phosphate broth and 10% fetal calf serum at 28°C.
Viral stocks of DENV-1 (Mochizuki strain), DENV-2 (New Guinea C strain), DENV-3 (H87 strain), and DENV-4 (H241 strain) were prepared as culture fluids from C6/36 cells infected with the individual serotypes and cultured for 7–9 days. In addition, the Nakayama strain of Japanese encephalitis virus was also similarly cultured in C6/36, and the culture fluid was used as a virus stock. Infectivity titers of these viruses were determined by the number of focus-forming units, as described previously.Citation25
Mouse immunization and monoclonal antibody preparation
For preparation of DENV-2 antigens, we used two types of antigens, ie, B7-cells infected with DENV-2 and 50% brain homogenate with phosphate-buffered solution (−) from suckling BALB/c mice that were injected with DENV-2. B7-cells infected with DENV-2, at a multiplicity of infection of 0.1 and cultured for 2 days, were harvested by scraping and precipitated by centrifugation at 1000 rpm for 5 minutes. Brain homogenate was prepared by intracerebral injection of DENV-2 in suckling mice. As soon as the injected mice showed symptoms, their brains were collected and frozen at −80°C until use. Antigens were kept at −80°C until use for immunization of mice. Three 4-week-old female BALB/c mice (National Laboratory Animal Center, Mahidol University, Bangkok, Thailand) were immunized with 2.5 × 106 infected cells or 300 μL of homogenized brain mixed with complete Freund’s adjuvant (Sigma-Aldrich, Saint Louis, MO), as described previously.Citation26 Each mouse was injected intraperitoneally with 300 μL of mixed antigen. Immunized mice were intraperitoneally boost-immunized 3–4 times with similarly prepared antigens without adjuvant. This study was approved by the Faculty of Tropical Medicine Animal Care and Use Committee, Mahidol University, Bangkok, Thailand (FTM-ACUC 2011/003). Three days after the final booster immunization, splenocytes were prepared and subjected to fusion with PAI cells using polyethylene glycol 1500 (Roche Diagnostic Corporation, Basel, Switzerland). Fused cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 15% fetal calf serum and hypoxanthine-aminopterin-thymidine (Gibco, Grand Island, NY). Monoclonal antibodies produced from hybridomas were screened with DENV-2-infected Vero cells by an immunofluorescence assay. Hybridomas were cloned twice by limiting dilutions using 96-well microplates.
Expression of DENV-2 capsid protein
The DENV-2 capsid protein expression plasmid, pCAGGS-PM2 FLAG-DEN2 core 100-HA, was kindly provided by Y Matsuura at the Research Institute for Microbial Diseases of Osaka University, Suita, Osaka, Japan. Expression of this plasmid was confirmed by Western blotting and immunofluorescence assays using an anti-Flag M2 monoclonal antibody (Sigma-Aldrich). The plasmid vector was transfected by Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for Vero cells.
Immunofluorescence assay
Vero cells were seeded into 96-well plates for preparation of DENV antigens in infected cells and DENV-2 capsid protein in transfected cells. After incubation for 16–24 hours, they were infected with each serotype of DENV or transfected with pCAGGS-PM2 FLAG-DEN2 core 100-HA plasmid. Two days after infection or transfection, the cells were fixed with 4% paraformaldehyde in phosphate-buffered solution for 30 minutes at room temperature. Vero cells infected with Japanese encephalitis virus were also similarly prepared. The fixed cells were permeabilized with 1% Triton X-100 in phosphate-buffered solution for 5 minutes at room temperature, and then incubated with hybridoma culture fluid for one hour. They were then washed three times with phosphate-buffered solution and further treated with Alexa Fluor® 488 goat anti-mouse IgG antibody (Invitrogen) at a dilution of 1:500 for 45 minutes. Finally, they were washed three times with phosphate-buffered solution prior to observation by fluorescence microscopy (IX71, Olympus, Tokyo, Japan).
Western blotting assay
DENV-infected Vero cells were dissolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer with beta-mercaptoethanol and heated at 100°C for 5 minutes. The samples were separated in 12% SDS-PAGE gel and transferred to a polyvinylidene fluoride membrane (Millipore Corporation, Bedford, MA). The membrane was incubated for 12 hours with antibody produced by the hybridoma clones and then with horseradish peroxidase-conjugated anti-mouse IgG (KPL, Washington, DC) for one hour. The reactive viral protein was visualized using an ECL WB detection agent (GE Healthcare, Buckinghamshire, UK).
Phylogenetic analysis of capsid, envelope, and nonstructural 1 proteins
All available sequences of DENV-1 to DENV-4, Japanese encephalitis virus, and West Nile virus were downloaded from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/protein) on February 7, 2012. The capsid, envelope, and nonstructural 1 amino acid sequences were extracted from these sequences using the results of a BLAST (Basic Local Alignment Search Tool) homology searchCitation27 against the corresponding proteins. These sequences were aligned using MAFFT version 6.705bCitation28 after removing redundant sequences. Phylogenetic trees were constructed using the neighbor-joining methodCitation29 with MEGA5.Citation30 All positions containing gaps and missing data were eliminated.
Results
Preparation of mouse monoclonal antibodies against DENV
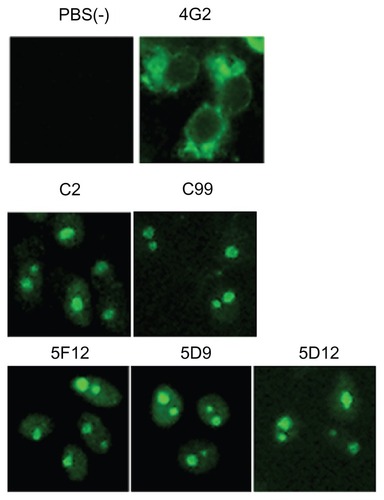
To prepare hybridoma clones producing anti-DENV monoclonal antibodies, spleen cells derived from mice immunized with DENV-2 infected cells were fused with myeloma cell lines as previously described.Citation26 After screening and single-cell cloning, we obtained a total of 60 hybridoma clones specifically reacted with DENV-2-infected Vero cells, but not with uninfected Vero cells, by indirect immunofluorescence assay. The 4G2 monoclonal antibody, available as an anti-flavivirus,Citation31 was used as a positive control for the immunofluorescence assay. This 4G2 reacted with the envelope protein of DENV in the cytoplasm of infected Vero cells (). Of the 60 clones generated in this study, five clones were reacted with the nuclei of infected cells (). Two clones (C2 and C99) were derived from mice immunized with the brain homogenate of BALB/c suckling mice injected with DENV-2, while three clones (5F12, 5D9, and 5D12) were derived from three mice immunized with B7-cells infected with DENV-2. All the other 55 monoclonal antibodies reacted with the cytoplasm of infected cells (data not shown).
Figure 1 Indirect immunofluorescence assay of anti-DENV-2 reacted with the nuclear protein of DENV-2 infected cells.
Monoclonal antibodies recognizing DENV capsid protein
To identify a viral protein recognized by five monoclonal antibodies which were shown to stain the nuclei of infected cells as above, a Western blot assay with the culture supernatants of individual hybridoma clones was carried out using the lysate of DENV-2-infected Vero cells that were treated with beta-mercaptoethanol and heated at 100°C. The C2 monoclonal antibody reacted specifically with 12 kDa DENV-2 capsid protein (). To confirm that this binding protein was the capsid protein, Vero cells were transfected with plasmid expressing recombinant DENV-2 capsid protein. This monoclonal antibody reacted with recombinant DENV-2 capsid protein in transfected cells on Western blotting and immunofluorescence assay ( and ). Although the other four monoclonal antibodies (C99, 5F12, 5D9, and 5D12) did not react with the 12 kDa capsid protein on Western blot assay (), these monoclonal antibodies similarly reacted with recombinant capsid protein in transfected Vero cells by immunofluorescence assay (). Thus, we concluded that these five monoclonal antibodies recognized the DENV capsid protein.
Figure 2 Identification of antigenic DENV-2 capsid protein. (A) Lysates of Vero cells infected with DENV-2 and 293T cells transfected with the DENV-2-C expression plasmid encoding Flag were used as antigens. Their reactivity against various antibodies was analyzed by Western blotting. Anti-Flag antibody was used as a positive control of capsid protein expression. Arrowheads showed 12 kDa. (B) Indirect immunofluorescence assay of monoclonal antibodies using Vero cells transfected with the DENV-2 capsid protein expression plasmid and fixed 48 hours following infection with 4% paraformaldehyde.
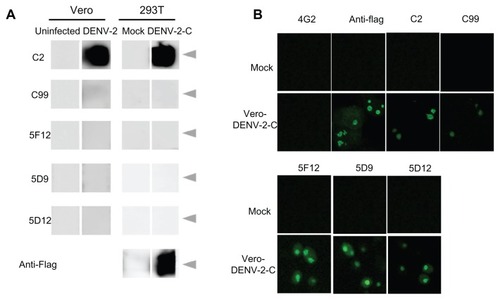
Table 1 Summary of cross-reactivity of MAb within Flavivirus and specificity MAb against DENV-2-C protein
Cross-reactivity of monoclonal antibodies to DENV capsid protein
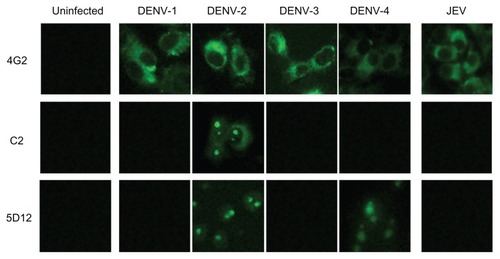
To determine the cross-reactivity of the five above-mentioned monoclonal antibodies to DENV capsid protein with each serotype of DENV and Japanese encephalitis virus, individual monoclonal antibodies in culture supernatants were examined by immunofluorescence assay using Vero cells infected with DENV-1 to DENV-4, as well as Japanese encephalitis virus (). The cross-reactivities of our five monoclonal antibodies are summarized in . Three monoclonal antibodies, C2, 5F12, and 5D9, were reacted specifically with only DENV-2 but not with the other serotypes, whereas the other two monoclonal antibodies, C99 and 5D12, were cross-reacted with DNEV-2 and DENV-4. None of these five monoclonal antibodies showed cross-reactivity with DENV-1 or DENV-3. In addition, there were no positive reactions with these five monoclonal antibodies in Vero cells infected with Japanese encephalitis virus ( and ).
Figure 3 Determination of cross-reactivities of monoclonal antibodies to flavivirus.
Phylogenetic analysis of flavivirus
Examination of anti-DENV capsid protein monoclonal antibodies for their cross-reactivity among four serotypes of DENV, as well as Japanese encephalitis virus and West Nile virus belonging to the same flavivirus, revealed only limited cross-reactivity. Therefore, we examined the situations for individual serotypes of DENV as well as Japanese encephalitis virus and West Nile virus by analysis of phylogenetic trees. The sequences of flavivirus capsid, envelope, and nonstructural 1 proteins all derived from the National Center For Biotechnology Information database were used for construction of individual phylogenetic trees. The numbers of the amino acid sequences used for phylogenetic analysis were shown in .
Table 2 Number of amino acid sequences of Flavivirus extracted from NCBI database
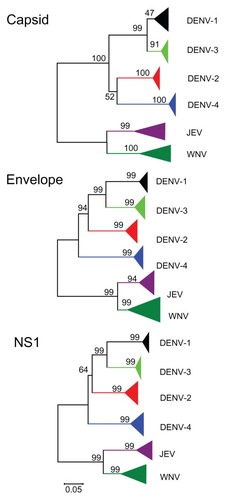
As observed in the cross-reactivity of monoclonal antibodies, phylogenetic analysis of the capsid protein showed that DENV-2 and DENV-4 were clustered in the same branch, while DENV-1 and DENV-3 were clustered in the other branch (, capsid protein). This meant that the capsid protein of DENV-2 was most closely related phylogenetically to that of DENV-4, but very far from those of DENV-1 and DENV-3. On the other hand, phylogenies of the envelope and nonstructural 1 proteins showed different branching orders among the four serotypes, with DENV-4 being the first diverge, followed by DENV-2, and the final diverge being between DENV-1 and DENV-3 (, envelope and nonstructural 1 proteins). The distances between DENV clusters seem to be comparable with those between Japanese encephalitis virus and West Nile virus (). Furthermore, clusters in the flavivirus capsid protein showed more diversity than those in the flavivirus envelope and nonstructural 1 proteins ().
Figure 4 Phylogenetic trees of flavivirus capsid, envelope, and nonstructural 1 protein inferred using the neighbor-joining method.
Discussion
In this study, we produced mouse monoclonal antibodies to the capsid protein of DENV. Five hybridoma cells producing anti-capsid monoclonal antibodies were successfully generated and characterized. Interestingly, none of the antibodies generated could bind with all serotypes of DENV. Consequently, this result was greatly different from our initial expectations. Phylogenetic tree analysis using database-derived sequences of DENV-1 to DENV-4 supported the above result, because capsid protein variations were higher than our initial expectation, as confirmed by comparison with other viral proteins, such as the envelope and nonstructural 1 proteins.
By immunization of BALB/c mice with DENV-2 antigens, we obtained only five monoclonal antibodies reactive with DENV capsid protein among the 60 monoclonal antibodies. Similarly, it has been reported that only a few anti-DENV capsid protein monoclonal antibodies are obtained by immunization with the whole virus.Citation32 In this study, the C2 monoclonal antibody reacted with DENV capsid protein by both Western blotting and immunofluorescence assays, while the C99, 5F12, 5D9, and 5D12 monoclonal antibodies reacted with the capsid protein only on immunofluorescence assay. These results suggest that the C2 monoclonal antibody recognized a linear epitope, and other monoclonal antibodies recognized conformational epitopes, and the structure of these conformation epitopes might be lost by cell-lysis buffer treatment. Further work is needed on the structural analysis of this epitope.
Three monoclonal antibodies were specific to DENV-2 and the other two clones were cross-reactive with DENV-2 and DENV-4. Previous research on anti-DENV capsid protein has reported similar results for serotype specificity and/or low cross-reactivity with DENV capsid protein.Citation6,Citation32–Citation34 However, the reason for the low cross-reactivity of DENV capsid protein has not been discussed in these papers. Our bioinformatics characterization of DENV capsid protein sequences from the database revealed that DENV-2 and DENV-4 were clustered in the same branch, while DENV-1 and DENV-3 clustered in the other branch (). However, these classifications of DENV capsid protein were not correlated with those of the DENV envelope and nonstructural 1 proteins. In general, epitopes recognized with antibodies are short amino acid sequences or protein conformations; therefore, it may be difficult to discuss serological cross-reactivity using only phylogenetic trees from amino acid sequences. However, our bioinformatics characterization results for the capsid protein may correlate well with the results for anti-capsid monoclonal antibodies obtained by immunization with DENV-2 antigens cross-reactive with DENV-2 and DENV-4, but not with those for DENV-1 and/or DENV-3. The phylogenetic tree for the flavivirus capsid proteins also showed that the distance between DENV-2 and DENV-4 clusters and between DENV-1 and DENV-3 clusters was comparable with that between Japanese encephalitis virus and West Nile virus. This bioinformatics information suggests that DENV-2 capsid antibodies that are cross-reactive with DENV-4 are relatively frequent, whereas those that are cross-reactive with DENV-1 and/or DENV-3 are rare (). The diversity between DENV serotypes was comparable with those between Japanese encephalitis virus and West Nile virus (). These tendencies were correlated with those not only in capsid proteins, but also in other proteins such as envelope and nonstructural 1 proteins. Our results show that phylogenetic distances between the four serotypes of DENV were as different as that of Japanese encephalitis virus and West Nile virus. However, the diversity of flavivirus capsid protein was much higher than that for envelope and nonstructural 1 proteins. Therefore, these data suggest that there may be difficulty in establishing monoclonal antibodies against DENV capsid protein that are cross-reactive with all the subtypes of DENV. The monoclonal antibodies specific for DENV-2 generated in this study could be used for serotyping of DENV infection. However, DENV capsid protein can be used for serotyping of DENV infection but not for rapid diagnostic test kits to detect all four DENV serotypes.
The levels of diversity among the DENV serotypes reflect the differences between species of flavivirus. This high diversity of DENV correlates with the classification of individual DENV serotypes. However, it is well known that there is significant diversity among genotypes for the individual DENV serotypes.Citation35,Citation36 These results suggest that individual DENV serotypes have evolved in their own particular way, unlike other flaviviruses.
In this study, the monoclonal antibodies generated specifically for DENV-2 capsid protein can be used for serotyping of DENV infection and for early diagnosis of high titers of viremia in patients with hemorrhagic Dengue fever or Dengue shock syndrome caused by DENV-2 infection. We could not obtain monoclonal antibodies cross-reactive with all four serotypes using mice immunized with a sole serotype of DENV. Our phylogenetic analysis of DENV and the high diversity of the DENV capsid protein sequence support this conclusion.
Acknowledgments
We are grateful to Pratap Singhasivanon (Faculty of Tropical Medicine, Mahidol University) and Shigeyuki Hamada (Research Collaboration Center on Emerging and Re-emerging Infections, Research Institute for Microbial Diseases, Osaka University) for their valuable help with this study. The manuscript was proofread by the Medical English Service (Kyoto, Japan). This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases program managed by the Ministry of Education, Cultures, Sports, Science, and Technology of Japan.
Disclosure
The authors report no conflicts of interest in this work.
References
- GublerDJEpidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st centuryTrends Microbiol200210210010311827812
- MackenzieJSGublerDJPetersenLREmerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue virusesNat Med200410Suppl 12S98S10915577938
- LindenbachDRRiceCMFlaviviridae: The Viruses and their ReplicationPhiladelphia, PALippincott Williams & Wilkins20014
- KuhnRJZhangWRossmannMGStructure of dengue virus: implications for flavivirus organization, maturation, and fusionCell2002108571772511893341
- SangiambutSKeelapangPAaskovJMultiple regions in dengue virus capsid protein contribute to nuclear localization during virus infectionJ Gen Virol200889Pt 51254126418420804
- BulichRAaskovJGNuclear localization of dengue 2 virus core protein detected with monoclonal antibodiesJ Gen Virol199273Pt 11299930031279106
- KeelapangPSriburiRSupasaSAlterations of pr-M cleavage and virus export in pr-M junction chimeric dengue virusesJ Virol20047852367238114963133
- AndersenJSLamYWLeungAKNucleolar proteome dynamicsNature20054337021778315635413
- BoisvertFMvan KoningsbruggenSNavascuesJLamondAIThe multifunctional nucleolusNat Rev Mol Cell Biol20078757458517519961
- OhWYangMRLeeEWJab1 mediates cytoplasmic localization and degradation of West Nile virus capsid proteinJ Biol Chem200628140301663017416882664
- MoriYOkabayashiTYamashitaTNuclear localization of Japanese encephalitis virus core protein enhances viral replicationJ Virol20057963448345815731239
- MoriishiKOkabayashiTNakaiKProteasome activator PA28 gamma-dependent nuclear retention and degradation of hepatitis C virus core proteinJ Virol20037719102371024912970408
- WestawayEGKhromykhAAKenneyMTMackenzieJMJonesMKProteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleusVirology1997234131419234944
- MuneneESongokENyamongoJALangatDKOtsyulaMEvaluation of HIV ELISA diagnostic kit developed at the Institute of Primate Research, Nairobi, KenyaAfr J Health Sci200293–411712217298154
- ChomelJJThouvenotDOnnoMKaiserCGourreauJMAymardMRapid diagnosis of influenza infection of NP antigen using an immunocapture ELISA testJ Virol Methods198925181912674180
- OttekenANickSBergterWIdentification of a gag protein epitope conserved among all four groups of primate immunodeficiency viruses by using monoclonal antibodiesJ Gen Virol199273Pt 10272127241383399
- MinassianAAKalyanaramanVSGalloRCPopovicMMonoclonal antibodies against human immunodeficiency virus (HIV) type 2 core proteins: cross-reactivity with HIV type 1 and simian immunodeficiency virusProc Natl Acad Sci U S A19888518693969432457921
- ZakiACoudrierDYousefAIFakeehMBouloyMBillecocqAProduction of monoclonal antibodies against Rift Valley fever virus: application for rapid diagnosis tests (virus detection and ELISA) in human seraJ Virol Methods20061311344016102851
- KimuraTRokuharaAMatsumotoANew enzyme immunoassay for detection of hepatitis B virus core antigen (HBcAg) and relation between levels of HBcAg and HBV DNAJ Clin Microbiol20034151901190612734224
- FabriziFBrombergJElliADixitVMartinPReview article: hepatitis C virus and calcineurin inhibition after renal transplantationAliment Pharmacol Ther200522865766616197487
- SaijoMNiikuraMIkegamiTKuraneIKurataTMorikawaSLaboratory diagnostic systems for Ebola and Marburg hemorrhagic fevers developed with recombinant proteinsClin Vaccine Immunol200613444445116603611
- DussartPLabeauBLagathuGEvaluation of an enzyme immunoassay for detection of dengue virus NS1 antigen in human serumClin Vaccine Immunol200613111185118916988003
- KumarasamyVWahabAHChuaSKEvaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infectionJ Virol Methods20071401–2757917140671
- KanaiYChittaganpitchMNakamuraIDistinct propagation efficiencies of H5N1 influenza virus Thai isolates in newly established murine respiratory region-derived cell clonesVirus Res2010153221822520709117
- KurosuTKhamlertCPhanthanawiboonSIkutaKAnantapreechaSHighly efficient rescue of dengue virus using a co-culture system with mosquito/mammalian cellsBiochem Biophys Res Commun2010394239840420214880
- MasrinoulPDiataMOPambudiSLimkittikulKIkutaKKurosuTHighly conserved region 141168 of the NS1 protein is a new common epitope region of dengue virusJpn J Infect Dis201164210911521519123
- AltschulSFMaddenTLSchafferAAGapped BLAST and PSI-BLAST: a new generation of protein database search programsNucleic Acids Res19972517338934029254694
- KatohKTohHImproved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based frameworkBMC Bioinformatics2008921218439255
- SaitouNNeiMThe neighbor-joining method: a new method for reconstructing phylogenetic treesMol Biol Evol1987444064253447015
- TamuraKPetersonDPetersonNStecherGNeiMKumarSMEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methodsMol Biol Evol201128102731273921546353
- FalconarAKIdentification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteinsArch Virol1999144122313233010664386
- PuttikhuntCOng-AjchaowlerdPPrommoolTProduction and characterization of anti-dengue capsid antibodies suggesting the N terminus region covering the first 20 amino acids of dengue virus capsid protein is predominantly immunogenic in miceArch Virol200915481211122119565324
- TadanoMMakinoYFukunagaTOkunoYFukaiKDetection of dengue 4 virus core protein in the nucleus. I. A monoclonal antibody to dengue 4 virus reacts with the antigen in the nucleus and cytoplasmJ Gen Virol198970Pt 6140914152471810
- GagnonSJZengWKuraneIEnnisFAIdentification of two epitopes on the dengue 4 virus capsid protein recognized by a serotype-specific and a panel of serotype-cross-reactive human CD4+ cytotoxic T-lymphocyte clonesJ Virol19967011411478523518
- Rico-HesseRMolecular evolution and distribution of dengue viruses type 1 and 2 in natureVirology199017424794932129562
- HolmesECTwiddySSThe origin, emergence and evolutionary genetics of dengue virusInfect Genet Evol200331192812797969