Abstract
Purpose
Imbalance in the microbiota, dysbiosis, has been identified in inflammatory bowel disease (IBD). We explored the fecal microbiota in pediatric patients with treatment-naïve IBD, non-IBD patients with gastrointestinal symptoms and healthy children, its relation to IBD subgroups, and treatment outcomes.
Patients and methods
Fecal samples were collected from 235 children below 18 years of age. Eighty children had Crohn’s disease (CD), 27 ulcerative colitis (UC), 3 IBD unclassified, 50 were non-IBD symptomatic patients, and 75 were healthy. The bacterial abundance of 54 predefined DNA markers was measured with a 16S rRNA DNA-based test using GA-Map™ technology at diagnosis and after therapy in IBD patients.
Results
Bacterial abundance was similarly reduced in IBD and non-IBD patients in 51 of 54 markers compared to healthy patients (P<0.001). Only Prevotella was more abundant in patients (P<0.01). IBD patients with ileocolitis or total colitis had more Ruminococcus gnavus (P=0.02) than patients with colonic CD or left-sided UC. CD patients with upper gastrointestinal manifestations had higher Veillonella abundance (P<0.01). IBD patients (58%) who received biologic therapy had lower baseline Firmicutes and Mycoplasma hominis abundance (P<0.01) than conventionally treated. High Proteobacteria abundance was associated with stricturing/penetrating CD, surgery (P<0.01), and nonmucosal healing (P<0.03). Low Faecalibacterium prausnitzii abundance was associated with prior antibiotic therapy (P=0.001), surgery (P=0.02), and nonmucosal healing (P<0.03). After therapy, IBD patients had unchanged dysbiosis.
Conclusion
Fecal microbiota profiles differentiated IBD and non-IBD symptomatic children from healthy children, but displayed similar dysbiosis in IBD and non-IBD symptomatic patients. Pretreatment fecal microbiota profiles may be of prognostic value and aid in treatment individualization in pediatric IBD as severe dysbiosis was associated with an extensive, complicated phenotype, biologic therapy, and nonmucosal healing. The dysbiosis persisted after therapy, regardless of treatments and mucosal healing.
Plain language summary
Studies have shown a disturbed gut bacterial composition in chronic inflammatory diseases such as inflammatory bowel disease (IBD) (Crohn’s disease and ulcerative colitis).
In children, it might be difficult to diagnose IBD. Symptoms are often nonspecific, such as abdominal pain and altered bowel movements.
Dr Olbjørn et al investigated whether the bacterial composition from stool samples can help to diagnose and treat IBD in children.
They used advanced DNA profiling to identify and quantify bacteria. They compared the bacterial composition in stool from children with IBD with healthy children and children with gastrointestinal symptoms but without inflammation.
The researchers report that the bacterial composition in patients with IBD was very different than in healthy children. The differences persisted after treatment.
The bacterial composition in patients with gastrointestinal symptoms but no inflammation was similarly disturbed as in IBD patients.
The degree of disturbances in the bacterial composition in children with IBD correlated with the disease course and later therapy. Patients with higher numbers of “bad” bacteria, such as Proteobacteria, were more likely to need aggressive treatment and surgery.
In children with IBD, testing the bacterial composition in the stool before treatment can help physicians in targeting and individualizing treatments.
Introduction
The pathogenesis of the inflammatory bowel diseases (IBD), Crohn’s disease (CD), and ulcerative colitis (UC) is not fully understood, but IBD is thought to occur due to an exaggerated immune response to luminal microbial contents in the gastrointestinal tract in genetically susceptible individuals.Citation1 A rising incidence of IBD, especially in the pediatric population, has been demonstrated, and the influence of environmental changes, including diet and gut microbiota on the disease pathogenesis, is increasingly recognized.Citation2,Citation3 The gut microbiota is thought to play an important role not only in IBD but also in functional gastrointestinal disorders such as irritable bowel syndrome, which may display similar symptoms representing a differential diagnosis to IBD.Citation4,Citation5 Studies of the gut microbiota in IBD and functional gastrointestinal disorders have shown an imbalance, dysbiosis, with compositional changes, including decreased bacterial diversity and abundance.Citation6–Citation8 The shift in the gut microbiota seems to be associated with a depletion of beneficial vs a relative increase of pro-inflammatory bacteria.Citation9,Citation10 The diagnostic and prognostic significance of fecal microbiota profiles in children with gastrointestinal symptoms and IBD is not fully explored.
We hypothesized that the fecal microbiota composition could be helpful in diagnosing pediatric IBD patients and in predicting their prognosis. We aimed to assess differences in the abundance of fecal microbiota in treatment-naïve pediatric IBD patients at the time of diagnosis compared to healthy controls and pediatric non-IBD patients with gastrointestinal symptoms. We further explored the value of microbiota abundance in differentiating between IBD phenotypes, subsequent need of biologic therapy, surgery and treatment outcomes, and whether the microbiota changes with therapy.
Patients and methods
IBD patients, non-IBD symptomatic patients, and healthy controls
Patients enrolled in the present study were recruited from the catchment areas of two university hospitals in three population-based prospective epidemiological studies of treatment-naïve pediatric IBD in South-Eastern Norway (IBSEN II),Citation11,Citation12 Early IBD (in preparation), and EU IBD Character.Citation13 The inclusion periods for these three multicenter trials were from 2005 to 2015, all with identical protocols and inclusion criteria. Pediatric patients under 18 years, referred during the inclusion periods and believed to have IBD based on symptoms, were included. IBD was diagnosed in accordance with the Porto criteria.Citation14 Patients who did not meet the diagnostic criteria for IBD were included as non-IBD symptomatic controls. These patients had a macroscopically and histologicallly normal mucosa and normal MRI examinations. Healthy children and adolescents between the age of 2 and 18 years and recruited during the period of 2013–2014 from the same catchment areas as the patients delivered fecal samples and were included as healthy controls. They had no chronic diseases, no IBD in the family, followed a normal diet (children on exclusion diets, gluten-free, cows milk protein-free, vegetarian/vegan, were excluded), had not traveled outside Europe or used antibiotics within the last 6 months, had no recorded gastrointestinal complaints, did not use proton pump inhibitors, and had normal fecal calprotectin levels (<50 mg/kg).Citation15
Clinical, endoscopic, radiological, and laboratory data
Age, gender, symptoms, disease activity index scores, disease, and family history of the IBD and non-IBD symptomatic patients were registered as previously described.Citation11,Citation12,Citation16 The Paris classification was used to characterize disease distribution and behavior.Citation17 In patients, feces were sampled at home in three designated containers without additives on the day before endoscopy, kept refrigerated or frozen, and brought to the hospital the next day. Fecal sample from one container was analyzed for calprotectin (FeCal-test, Bühlmann, Basel, Switzerland), the second for pathogenic bacteria, and the third container with feces was frozen at −80°C for later microbiota analysis. The healthy controls received two designated fecal sampling kits at home for handling of samples before deliverance to Genetic Analysis AS, Oslo, Norway. One sample was analyzed for fecal calprotectin (FeCal-test, Bühlmann); the other was frozen at −80°C and stored for later microbiota analysis. For all samples, the maximum time interval until frozen at −80°C was 3 days; thereafter the samples were kept frozen and not thawed until analysis.
Microbiota analysis
The microbiota was analyzed using the GA-Map™ technology (Genetic Analysis AS), a PCR, and 16S RNA-based analysis. The method uses a targeted approach to detect predefined bacteria believed to be important in identifying and characterizing gut bacteria dysbiosis.Citation18 The test measures relative bacterial abundance based on the fluorescence signal strength (FSS) of bacterial DNA markers. The markers are targeting variable regions V3–V7 of the bacterial 16S rRNA gene. The method utilizes 54 bacterial markers (Table S1), covering more than 300 bacteria at different taxonomic levels: 26 species specific, 19 detect genus specific, and 9 bacteria at higher taxonomic levels (phyla, class, and family). All samples were analyzed at the same time point. The laboratory was blinded for the diagnosis of IBD, non-IBD, or healthy.
IBD treatment
Treatment was decided individually, prospectively, at the discretion of the treating pediatrician. Initial treatment options to induce remission were exclusive enteral nutrition in CD and corticosteroids and/or 5-aminosalicylic acids in CD and UC patients. Maintenance therapy with azathioprine or methotrexate was in general started simultaneously (). The indication for surgery or treatment with biologic therapy (TNF blockers) was failure to induce remission with conventional treatments or relapse after primary induction.
Table 1 Disease extent and behavior at diagnosis according to the Paris classification and treatments in pediatric IBD patients
Statistical analyses
Data were described using counts and percentages for categorical data and medians and ranges for continuous data. To explore the ability of all 54 bacterial markers to distinguish between IBD, non-IBD symptomatic patients, and healthy controls, we performed principal component analysis. The FSSs from the 54 markers were added for each patient, and the sum illustrated a relative abundance, denoted the total signal strength. Crude comparisons between groups were performed using Mann–Whitney Wilcoxon tests and Wilcoxon signed-rank tests (before and after treatment) for continuous variables and chi-squared tests for categorical data.
Areas under the curves were calculated and receiver operating characteristic analysis conducted to evaluate the performance of selected bacterial abundances in distinguishing IBD phenotypes and treatments. All tests were two-sided. P-values <0.05 were considered statistically significant. We regarded our study exploratory; therefore, we did not correct for multiple testing. However, in order to validate our results, each observation was randomized into a test set or a training set so that the number of observations was equal in both sets. Only the statistically significant differences confirmed in the training set are reported. All analyses were performed using SPSS, statistical software, version 24 (SPSS Inc., Chicago, IL, USA) and Stata, version 9.
Ethical considerations
The study was conducted with informed patients and parental/guardian written consent as appropriate and with full ethical approval, in accordance with the Declaration of Helsinki, and with approval by the Regional Committee for Medical Research Ethics, South-Eastern Norway, reference no. REK S-04209.
Results
Of the 235 included children and adolescents, IBD was diagnosed in 110 patients (80 CD, 27 UC, and 3 IBDU) (), 50 patients were included as non-IBD symptomatic patients, and 75 healthy children served as controls. None of the non-IBD symptomatic patients have developed IBD as of December 1, 2018. IBD, non-IBD, and healthy controls were comparable concerning all demographic variables except for more females among the non-IBD patients and a slightly lower median age in the healthy controls ().
Table 2 Demographics and laboratory tests of IBD, non-IBD patients, and healthy controls at baseline
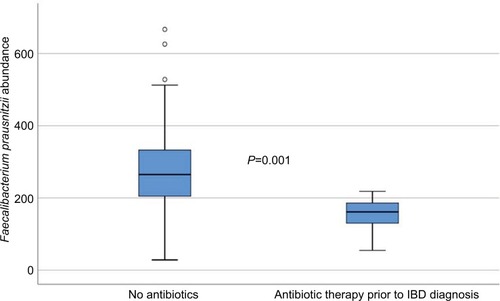
The bacterial abundances were compared between the three pediatric groups, healthy controls, IBD patients and non-IBD symptomatic patients, as well as between subgroups of IBD and after treatment in 31 of the IBD patients. To investigate the impact of antibiotics on microbiota profiles of the IBD patients, they were grouped according to whether they had received antibiotics within 3 months prior to diagnosis or not, and analyzed separately. Eight of the 110 IBD patients had received antibiotics, and these patients had significantly lower abundance of Faecalibacterium prausnitzii (P=0.001) compared to IBD patients without prior antibiotic therapy (). However, excluding these patients from the statistical analyses did not impact the other results presented in the material.
Figure 1 Faecalibacterium prausnitzii abundance in IBD patients according to whether they had received antibiotics prior to the diagnosis (measured in fluorescence signal strength in 1,000 units).
Microbiota in relation to age
We found significant differences in microbiota abundance when comparing healthy children below (n=38) and above (n=37) 10 years of age. Healthy children aged <10 years had lower abundance of Clostridiales and higher abundance of Bifidobacterium, both P<0.01. These differences were not replicated in the patients as we did not find any differences in bacterial profiles between high and low age groups in the IBD and non-IBD symptomatic patient groups. Additional post hoc analysis with an age matched selection of controls did not influence outcome/differences between patients and healthy.
Microbiota in IBD and non-IBD vs healthy
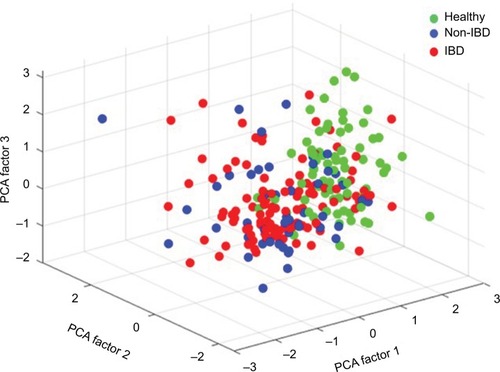
In all symptomatic patients, regardless of IBD or non-IBD status, the total signal strength, measured as the sum of the 54 FSSs, was significantly lower compared to healthy controls, illustrating that the patients had lower abundance of the predefined bacterial markers. Patients had reduced bacterial abundances in 51/54 markers, P<0.001 (). The only bacterial marker that was more abundant in patients (IBD and non-IBD) compared to healthy controls was Prevotella (P<0.01). The abundances of Lachnospiraceae and Bacteroides were similar in all groups. The principal component analysis plot visualizes how the microbiota composition differs between IBD, non-IBD, and healthy and overlaps between IBD and non-IBD symptomatic patients ().
Figure 2 Boxplot illustrating the differences in the total fluorescence signal strength measured in 1,000 units between IBD, non-IBD symptomatic patients, and healthy controls.
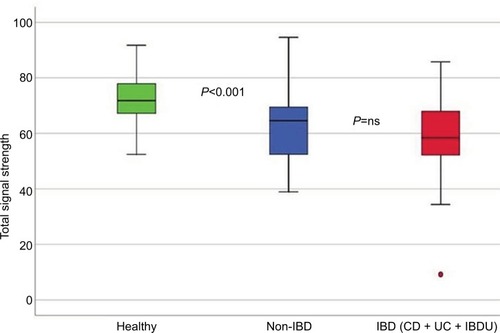
Figure 3 Principal component analysis, illustrating the difference in microbiota abundance of all 54 bacterial probes between IBD, non-IBD symptomatic patients, and healthy controls.
Abbreviations: IBD, inflammatory bowel disease; PCA, principal component analysis.
Microbiota in IBD vs non-IBD
The bacterial abundances were similarly dysbiotic in IBD and non-IBD symptomatic patients; however, one marker targeting the Firmicutes phylum was significantly less abundant in IBD patients compared to non-IBD patients (P<0.01), as well as Eubacterium rectale (P<0.01), Eubacterium biforme/Streptococcus agalactiae (P=0.04), Parabacteroides, and Bifidobacterium species (both P=0.02).
Microbiota in IBD patients
The fecal microbiota abundances did not differ between UC and CD, except that CD patients had lower abundance of Mycoplasma hominis (P<0.02).
Microbiota related to disease distribution and behavior in IBD patients
IBD patients with extensive disease, ileocolitis in CD or extensive colitis in UC, had higher abundance of Rumi-nococcus gnavus (P=0.02) compared to CD patients with isolated colonic disease and UC patients with limited disease distribution (left-sided colitis or proctitis). CD patients with upper gastrointestinal involvement had higher Veillonella abundance (P<0.01) compared to patients without upper gastrointestinal lesions.
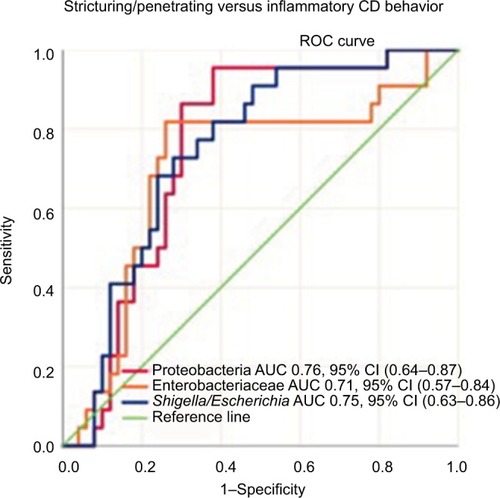
CD patients with a high abundance of Proteobacteria were more likely to have complicated disease behavior, stricturing or penetrating disease, compared to patients with lower levels of these bacteria, P<0.01 ().
Figure 4 Sensitivity and specificity of Proteobacteria, Enterobacteriaceae, and Shigella/Escherichia abundance in differentiating Crohn’s disease phenotypes (stricturing/penetrating vs inflammatory disease behavior) using the area under the receiver operating characteristics curve analysis.
Microbiota and association with treatment
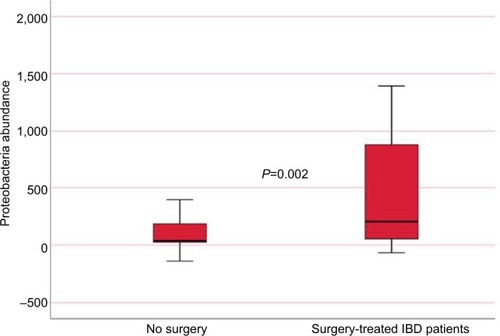
IBD patients who were treated with biologic therapy, 64 (58%), had lower abundance of Firmicutes (P=0.015) and M. hominis (P=0.009) compared to conventional treated patients (). Seventeen (15%) of the IBD patients required surgery, and mucosal healing (assessed by ileocolonoscopy) was not achieved in 40 (36%) of the patients despite medical therapy. Surgery and lack of mucosal healing were associated with higher abundance of Proteobacteria (P=0.002 and P=0.011) () and lower baseline abundance of F. prausnitzii (P=0.02 and P=0.017), respectively, compared to nonoperated IBD patients and patients with mucosal healing.
Figure 5 Sensitivity and specificity of Firmicutes and Mycoplasma hominis abundance in differentiating conventional- vs biologic therapy-treated IBD patients using the area under the receiver operating characteristics curve analysis.
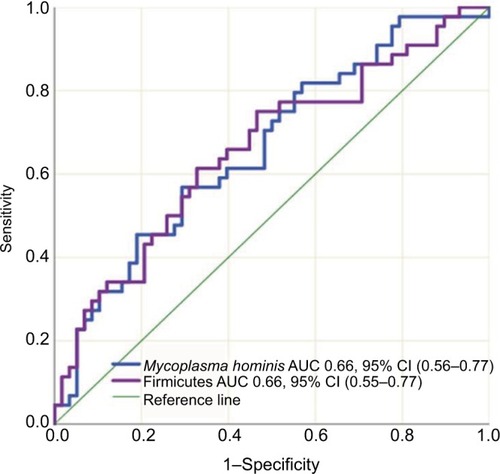
Figure 6 Proteobacteria abundance in IBD patients according to whether they needed surgery or not (measured in fluorescence signal strength in 1,000 units).
Of the IBD patients (22 CD and 9 UC) with repeated microbiota analysis at follow-up 18 months after treatment, 15 (48%) patients had received biologic therapy and 18 (58%) were in remission with mucosal healing. The microbiota composition and bacterial profiles were unchanged for 53 of 54 markers after treatment, regardless of treatment modality received and remission status. One marker targeting Eubacterium hallii species was less abundant after treatment, P=0.03.
Microbiota and association with fecal calprotectin
IBD patients with fecal calprotectin levels above 1,000 mg/kg (31 CD, 12 UC) had significantly higher abundance of Proteobacteria (P=0.012) and Prevotella (P=0.011) than patients with lower levels (<1,000 mg/kg) of fecal calprotectin (). Fecal calprotectin over 1,000 was associated with subsequent biologic therapy, P=0.001, but not with later surgery.
Discussion
In the present prospective study of newly diagnosed children and adolescents with IBD, we demonstrated dysbiosis in both treatment-naïve pediatric IBD and non-IBD symptomatic patients. Their fecal microbiota differed significantly from the microbiota of healthy children with lower bacterial abundances measured with the GA-Map technology. Our non-IBD symptomatic patients consisted of pediatric patients admitted to the hospital due to symptoms and findings suspicious of IBD, but without evidence of inflammation during workup. Some of these patients may have had preclinical/latent IBD or other conditions such as disturbed permeability and motility influencing the study results. We believe most of these non-IBD symptomatic patients have functional gastrointestinal disorders. Ideally we should have further characterized and subtyped these patients with the use of Rome criteria for functional gastrointestinal disorders. However, due to the limited sample size of 50 non-IBD symptomatic patients, further subclassification would reduce the statistical power to reveal clinical significant differences between the groups.
There was a similar dysbiotic profile with reduced microbial abundance in IBD and non-IBD compared to healthy individuals in the present study; thus the bacterial profiles provided by the GA-Map technology performed less well than fecal calprotectin in detecting inflammation and discriminating IBD from non-IBD symptomatic patients. However, the finding of dysbiosis in non-IBD symptomatic patients may confirm the relevance of their symptoms and discomfort. Presence and characterization of dysbiosis enables the physician to diagnose “functional” disease in a positive manner.
Within the group of patients diagnosed with IBD, we found that bacterial abundances at baseline seemed to be associated with disease extension, phenotype, biologic therapy, surgery, and mucosal healing. At follow-up, after treatment, the dysbiosis was still present and its status mainly unchanged in IBD patients.
We found reduced abundances of beneficial Eubacterium and Bifidobacterium species in IBD and non-IBD symptomatic patients compared to healthy children, in agreement with previous adultCitation6,Citation19,Citation20 and pediatric studies.Citation21–Citation23 Eubacteria and Bifidobacterium are known to inhibit the growth of potentially pathogenic speciesCitation24 and produce short-chain fatty acids (SCFAs) through fermentation of dietary fiber. SCFAs are important energy sources for enterocytes and contribute to homeostasis of colonic regulatory T cell populations.Citation25 The reduction of protective commensal microbes and concomitant loss of their protective function can have an influence on development of IBD and the disease course. As expected, Bifidobacterium was more abundant in healthy children below 10 years of age than in the healthy adolescents.Citation26 We found no difference in bacterial abundance between age groups for our IBD and non-IBD symptomatic patients. This may be due to disease state being a stronger driver of the microbiota composition than age.
Patients with IBD have an expansion of pro-inflammatory bacteria such as Prevotella,Citation27,Citation28 R. gnavus,Citation29 and Veillonella.Citation22,Citation28 Veillonella was enriched in our CD patients with upper gastrointestinal involvement. R. gnavus, a bacterium that expresses beta-glucuronidase activity, which may cause local inflammation, was associated with more extensive IBD distribution in our patients. Prevotella, R. gnavus, and Proteobacteria have been found to correlate with markers of disease activity and inflammation,Citation28,Citation30 which were reproduced in the present study. Proteobacteria are pathobionts, meaning that they may expand as a result of a microbial imbalance and exert pathogenic effects on the host and are consistently reported enriched in IBD.Citation31–Citation33 Our CD patients with a complicated phenotype had high abundance of Proteobacteria, in accordance with the previous reports. Proteobacteria enrichment has been associated with early relapse after induction of remission with exclusive enteral nutrition in pediatric CD,Citation34 and in our patients, high abundance was associated with the need for surgery and lack of mucosal healing. These findings implicate that Proteobacteria abundance might be a marker for an aggressive disease course with a higher risk of treatment failure.
F. prausnitzii, a highly abundant human gut microbe, is reported to be reduced in both adult and pediatric patients with IBD.Citation6,Citation13,Citation35,Citation36 It acts as a protective factor for the intestinal mucosa, enhances barrier function, and can exert anti-inflammatory effects.Citation20,Citation37,Citation38 Our IBD patients who needed surgery and who did not achieve mucosal healing with therapy, as well as patients treated with antibiotics before the IBD diagnosis, had the lowest abundance of F. prausnitzii. This is in line with observations that low abundance of F. prausnitzii may predict nonresponse to anti-TNF therapy in UCCitation39 and relapse after infliximab termination in CD patients.Citation40 Studies have found baseline microbiota to be associated with treatment responses,Citation34,Citation36,Citation39 but how the microbiota composition and abundances change with treatment is less studied. The IBD patients in our sample with repeated fecal microbiota analysis displayed persistent, unchanged dysbiosis after treatment, regardless of treatment modalities and remission status. Similar results have been reported in another pediatric study, where the dysbiosis improved, but nonetheless persisted despite mucosal healing.Citation41 Lewis et al found that effective exclusive enteral nutrition and TNF blocker therapy reduced but failed to eliminate the dysbiosis of pediatric CD patients.Citation42 Others have found the fecal microbiota to become more dysbiotic with dietary treatment such as exclusive enteral nutrition.Citation43–Citation45 Perhaps sustained and deep remission requires normalization of the gut dysbiosis, or maybe it is not possible to reverse the dysbiosis once the gut homeostasis is perturbed as fundamentally as it is in IBD. Measuring relative fecal microbiota abundance might not be an optimal method as it is not suited to determine the effects of dysbiosis, giving no information about the functional consequences. As a prognostic tool, fecal microbiota profiles may still be of value, also in established IBD patients on treatment, as the dysbiosis remained despite treatment and remission. However, due to the small number of patients with repeated sampling, firm conclusions cannot be drawn.
Regarding fecal microbial differences between CD and UC, the literature has been conflicting. Similarly, as in our report, some previous studies did not find major differences in bacterial profiles between active CD and active UC.Citation23,Citation36
The strength of our study is the extensive workup, characterization, and classification of our IBD patients. All non-IBD symptomatic patients underwent the same procedures as the IBD patients. Upper and lower endoscopies as well as MRI of the small intestine were performed, and for patients included in the IBSEN II cohort these investigations were repeated after 1–2 years of follow-up. The fact that none of the non-IBD symptomatic patients have been diagnosed with IBD despite several (minimum 3, maximum 13) years of follow-up makes misclassifications and undiagnosed IBD less likely.
The healthy controls were not investigated in the same manner as the patients, as invasive tests in healthy children are considered unethical. Even though children with gastrointestinal complaints, recent antibiotic exposure, and elevated fecal calprotectin were excluded as healthy controls, some could have had conditions that may have influenced the study results, as there is substantial evidence that diseases outside of the gastrointestinal tract influence the gut microbiota.Citation46
Dietary patterns and smoking are known to influence the microbiota;Citation45 therefore, we excluded patients on exclusion diets. None of our adolescents admitted to smoking.Citation47
The selection of microbes in the GA-map™ technology is based on literature studies and contain gut bacteria whose profiles are known to define dysbiosis in adults, with the inherent risk of not including bacteria that could be important in children and adolescents. Bacterial 16S sequencing of all microbes would give additional results, but is more expensive. The same is true for shotgun metagenomic sequencing, encompassing all DNA of bacteria, viruses, and fungi. Together with an altered bacterial composition, studies have revealed that IBD patients have fungal dysbiosis as well as alterations in the intestinal virome, which we have not investigated in our study.Citation48,Citation49 Deep sequencing and shotgun metagenomic sequencing methods need bioinformatics tools and reference datasets that are still under development and not yet readily available for clinical practice. The GA-Map technology provided us with a commercially available and clinically validated (in adults) tool.
Our study has several limitations. First, the sample size is limited, reducing the statistical power to detect differences in microbiota composition as statistically significant. We did not adjust for multiple testing as we considered this study to be exploratory, increasing the risk for accepting false-positive associations. However, we validated our results by splitting our data into a training and a test set, and most associations estimated in the whole cohort remained statistically significant. The positive relationship between inflammation, increased abundance of pathobionts and concomitant loss of beneficial bacteria, is reassuring as it is in line with previous research reports.Citation50
Another limitation is the difference in storage time of the fecal samples, which may have influenced outcomes. Also, theoretically, the representativeness of the samples could have deteriorated during the timespan from collection until frozen. Based on previous experience and in vitro examinations,Citation18 the microbial material collected in different cohorts was not considered to be affected. Since repeated thawing is known to influence the microbiota, the samples were kept frozen until analysis.
We acknowledge that the GA-Map technology test measures the abundance of bacteria without giving information about the functional importance and highly abundant bacteria might not be functionally active.Citation51 Additionally, in the present study, we explored the fecal microbiota only. One study comparing mucosal associated microbiota with fecal microbiota reported that the ileal mucosa followed by the rectal mucosa obtained the best performance in classifying CD and that stool samples performed less well.Citation22 Mucosa associated microbiota must be sampled by invasive methods. In this study however, we wanted noninvasive methods to associate microbiota with disease state. Our findings show promise for microbiota profiles and abundance to be used in conjuncture with other prognostic factors and known biomarkers in an attempt to risk stratify and individualize treatments in pediatric IBD.
Conclusion
Fecal microbiota profiles similarly differentiated IBD and non-IBD symptomatic children from healthy children. Microbiota profiles with relative enrichment of Proteobacteria and low abundance of F. prausnitzii in newly diagnosed pediatric IBD seem to be associated with complicated disease phenotypes, subsequent need of biologic therapy, surgery, and nonmucosal healing. The dysbiosis persisted after therapy, regardless of treatments and remission status. The relative abundances of selected bacteria might be of value as prognostic markers in stratifying pediatric IBD into subgroups and aid in patient selection for early aggressive therapy in an effort to prevent a complicated disease course.
Author contributions
All authors have made substantial contributions in conception and study design, acquisition of data, or analysis and interpretation of data and taken part in drafting the article or revising it critically for important intellectual content. All authors have seen and approved the final version of the manuscript submitted to the journal and agree to be accountable for all aspects of the work. CO and GP contributed in planning the study, collecting data, analyzing, and interpreting the results and drafting the article. MCS, BN, ETE, and MHV contributed in planning the study, analyzing and interpreting the results, and drafting the article.
Acknowledgments
The authors thank Genetic Analysis AS, Oslo, Norway for conducting the analyses. A special thanks to all patients and parents who contributed to the study, to our colleagues and collaborators who were of assistance in collecting and analyzing the materials, and to the IBD Character Consortium for contributions in study conception and design: Jack Satsangi, Rahul Kalla, Alex T Adams, Elaine R Nimmo, Hazel E Drummond, Ray K Boyapati, Nicholas T Ventham, Nicholas A Kennedy, David C Wilson, Charles W Lees, Colin L Noble, Ian D Arnott, Gwo-Tzer Ho, Alan G Shand, Kate R O’Leary, Jørgen Jahnsen, Morten H Vatn, Tone M Tannæs, Aina EF Moen, Petr Ricanek, Simen Vatn, Christine Olbjørn, Trond Espen Detlie, Jonas C Lindstrom, Anna Frengen, Panpan You, Janne Sølvernes, Fredrik A Dahl, Gunn S Ekeland, Åsa V Keita, Johan D Söderholm, Henrik Hjortswang, Jonas Halfvarson, Daniel Bergemalm, Fernando Gomollón, Mauro D’Amato, Leif Törkvist, Christina Casén, Magdalena K Karlsson, Fredrik Hjelm, Mats Gullberg, Niklas Nordberg, Anette Ocklind, Erik Pettersson, Daniel Ekman, Mikael Sundell, Eddie Modig, Anne-Clémance Veillard, Renaud Schoemans, Dominique Poncelet, Céline Sabatel, Ivo G Gut, Marta Gut, Simon Heath, Monica Bayes, Angelika Merkel, and Ferdinando Bonfiglio. The study was partly funded by the EU FP7-Health-2012 project, IBD Character. Grant agreement no: 305676 (http://cordis.europa.eu/project/rcn/106191_en.html).
Supplementary material
Table S1 List of phyla and bacterial names of the GA-Map™ technology markers
Disclosure
Christine Olbjørn is a member of the advisory board of AbbVie and has received speaker honoraria from AbbVie, Nutricia, Norgine, Tillotts Pharma, and Mead Johnson. Morten H Vatn has been an advisor for Genetic Analysis and organizer of the International Advisory Board of Genetic Analysis, a member of the advisory board for Tillotts Pharma, and has received speaker honoraria from AstraZeneca, AbbVie, MSD, and Falk. Gøri Perminow is a member of the advisory board of AbbVie and is a member of the steering committee in the IBSEN III study. The IBSEN III study has received an Investigator Initiated Research Grant from Takeda and nonrestricted research grants from Ferring Pharmaceuticals and Tillotts Pharma. Christina Casén and Magdalena K Karlsson are employed by Genetic Analysis. The authors report no other conflicts of interest in this work.
References
- ChuHKhosraviAKusumawardhaniIPGene-microbiota interactions contribute to the pathogenesis of inflammatory bowel diseaseScience201635262891116112027230380
- NgSCShiHYHamidiNWorldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studiesLancet2018390101142769277829050646
- KaplanGGNgSCUnderstanding and preventing the global increase of inflammatory bowel diseaseGastroenterology2017152231332127793607
- LinLZhangJRole of intestinal microbiota and metabolites on gut homeostasis and human diseasesBMC Immunol2017181228061847
- SpillerRMajorGIBS and IBD – separate entities or on a spectrum?Nat Rev Gastroenterol Hepatol2016131061362127667579
- SartorRBWuGDRoles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approachesGastroenterology2017152232733927769810
- ChangCLinHDysbiosis in gastrointestinal disordersBest Pract Res Clin Gastroenterol201630131527048892
- SundinJOhmanLSimrenMUnderstanding the gut microbiota in inflammatory and functional gastrointestinal diseasesPsychosom Med2017
- MiyoshiJChangEBThe gut microbiota and inflammatory bowel diseasesTransl Res2017179384827371886
- HoldGLSmithMGrangeCWattEREl-OmarEMMukhopadhyaIRole of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years?World J Gastroenterol20142051192121024574795
- PerminowGBrackmannSLyckanderLGA characterization in childhood inflammatory bowel disease, a new population-based inception cohort from South-Eastern Norway, 2005-07, showing increased incidence in Crohn’s diseaseScand J Gastroenterol200944444645619117240
- OlbjørnCCvancarova SmåstuenMThiis-EvensenENakstadBVatnMHPerminowGSerological markers in diagnosis of pediatric inflammatory bowel disease and as predictors for early tumor necrosis factor blocker therapyScand J Gastroenterol201752441441927887202
- RicanekPVatnSKallaRMicrobiota alterations in treatment naïve IBD and non-IBD patients – the EU IBD character projectUnited Eur Gastroenterol J201645 supplA721A754
- IBD Working Group of the European Society for Paediatric Gastroenterology, Hepatology and NutritionInflammatory bowel disease in children and adolescents: recommendations for diagnosis – the Porto criteriaJ Pediatr Gastroenterol Nutr20054111715990620
- FagerbergULLööfLMerzougRDHanssonLOFinkelYFecal calprotectin levels in healthy children studied with an improved assayJ Pediatr Gastroenterol Nutr200337446847214508218
- OlbjørnCNakstadBSmåstuenMCThiis-EvensenEVatnMHPerminowGEarly anti-TNF treatment in pediatric Crohn’s disease. Predictors of clinical outcome in a population-based cohort of newly diagnosed patientsScand J Gastroenterol201449121425143125310799
- LevineAGriffithsAMarkowitzJPediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classificationInflamm Bowel Dis20111761314132121560194
- CasénCVebøHCSekeljaMDeviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBDAliment Pharmacol Ther2015421718325973666
- LopetusoLRPetitoVGrazianiCGut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: time for microbial marker of gastrointestinal disordersDig Dis2018361566528683448
- BennetSMOhmanLSimrenMGut microbiota as potential orchestrators of irritable bowel syndromeGut Liver20159331833125918261
- MaukonenJKolhoKLPaaselaMAltered fecal microbiota in paediatric inflammatory bowel diseaseJ Crohns Colitis20159121088109526351391
- GeversDKugathasanSDensonLAThe treatment-naive microbiome in new-onset Crohn’s diseaseCell Host Microbe201415338239224629344
- PapaEDocktorMSmillieCNon-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel diseasePLoS ONE201276e3924222768065
- SatokariRContentious host–microbiota relationship in inflammatory bowel disease – can foes become friends again?Scand J Gastroenterol2015501344225523554
- SmithPMHowittMRPanikovNThe microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasisScience2013341614556957323828891
- ArrietaM-CStiemsmaLTAmenyogbeNBrownEMFinlayBThe intestinal microbiome in early life: health and diseaseFront Immunol20145Suppl 142725250028
- ForbesJDvan DomselaarGBernsteinCNThe gut microbiota in immune-mediated inflammatory diseasesFront Microbiol2016719032108127462309
- MottaweaWChiangCKMühlbauerMAltered intestinal microbiotahost mitochondria crosstalk in new onset Crohn’s diseaseNat Commun2016711341927876802
- JoossensMHuysGCnockaertMDysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relativesGut201160563163721209126
- BerryDReinischWIntestinal microbiota: a source of novel biomarkers in inflammatory bowel diseases?Best Pract Res Clin Gastroenterol2013271475823768552
- FrankDNSt AmandALFeldmanRABoedekerECHarpazNPaceNRMolecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseasesProc Natl Acad Sci USA200710434137801378517699621
- MukhopadhyaIHansenREl-OmarEMHoldGLIBD – what role do Proteobacteria play?Nat Rev Gastroenterol Hepatol20129421923022349170
- KaakoushNODayASHuinaoKDMicrobial dysbiosis in pediatric patients with Crohn’s diseaseJ Clin Microbiol201250103258326622837318
- DunnKAMoore-ConnorsJMacintyreBEarly changes in microbial community structure are associated with sustained remission after nutritional treatment of pediatric Crohn’s diseaseInflamm Bowel Dis201622122853286227805918
- ThorkildsenLTNwosuFCAvershinaEDominant fecal microbiota in newly diagnosed untreated inflammatory bowel disease patientsGastroenterol Res Pract20132013170113
- KolhoKLKorpelaKJaakkolaTFecal microbiota in pediatric inflammatory bowel disease and its relation to inflammationAm J Gastroenterol2015110692193025986361
- MccarvilleJLCamineroAVerduEFNovel perspectives on therapeutic modulation of the gut microbiotaTherap Adv Gastroenterol201694580593
- BurmanSHoedtECPottengerSMohd-NajmanNSÓ CuívPMorrisonMAn (anti)-inflammatory microbiota: defining the role in inflammatory bowel disease?Dig Dis2016341–2647126982568
- MagnussonMKStridHSapnaraMAnti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota compositionJ Crohns Colitis201610894395226896085
- RajcaSGrondinVLouisEAlterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s diseaseInflamm Bowel Dis201420697898624788220
- ShawKABerthaMHofmeklerTDysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel diseaseGenome Med2016817527412252
- LewisJDChenEZBaldassanoRNInflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s diseaseCell Host Microbe201518448950026468751
- GerasimidisKBertzMHanskeLDecline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutritionInflamm Bowel Dis201420586187124651582
- MaclellanAConnorsJGrantSCahillLLangilleMvan LimbergenJThe Impact of Exclusive Enteral Nutrition (EEN) on the gut microbiome in Crohn’s disease: a reviewNutrients2017950447
- QiaoYQCaiCWRanZHTherapeutic modulation of gut microbiota in inflammatory bowel disease: more questions to be answeredJ Dig Dis2016171280081027743467
- GaufinTTobinNHAldrovandiGMThe importance of the microbiome in pediatrics and pediatric infectious diseasesCurr Opin Pediatr201830111712429206649
- LaneERZismanTLSuskindDLThe microbiota in inflammatory bowel disease: current and therapeutic insightsJ Inflamm Res201710637328652796
- SokolHLeducqVAschardHFungal microbiota dysbiosis in IBDGut20176661039104826843508
- NormanJMHandleySABaldridgeMTDisease-specific alterations in the enteric virome in inflammatory bowel diseaseCell2015160344746025619688
- NiJWuGDAlbenbergLTomovVTGut microbiota and IBD: causation or correlation?Nat Rev Gastroenterol Hepatol2017141057358428743984
- MoenAETannæsTMVatnSSimultaneous purification of DNA and RNA from microbiota in a single colonic mucosal biopsyBMC Res Notes20169132827352784
