?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
Barrett’s esophagus (BE), a metaplastic premalignant disorder, represents the primary risk factor for the development of esophageal adenocarcinoma. Chronic gastroesophageal reflux disease and central obesity have been associated with BE and esophageal adenocarcinoma, but relatively little is known about the specific genes that confer susceptibility to BE carcinogenesis.
Methods
A total of 74 patients with BE and 67 controls coming from six gastrointestinal Italian units were evaluated for six polymorphisms in four genes: XPC, XPD nucleotide excision repair (NER) genes, XRCC1 (BER gene), and glutathione S-transferase P1. Smoking status was analyzed together with the genetic data. Statistical analysis was performed through Artificial Neural Networks.
Results
Distributions of sex, smoking history, and polymorphisms among BE cases and controls did not show statistically significant differences. The r-value from linear correlation allowed us to identify possible protective factors as well as possible risk factors. The application of advanced intelligent systems allowed for the selection of a subgroup of nine variables. Artificial Neural Networks applied on the final data set reached mean global accuracy of 60%, reaching as high as 65.88%.
Conclusion
We report here results from an exploratory study. Results from this study failed to find an association among the tested single nucleotide polymorphisms and BE phenotype through classical statistical methods. On the contrary, advanced intelligent systems are really able to handle the disease complexity, not treating the data with reductionist approaches unable to detect multiple genes of smaller effect in predisposing to the disease.
Impact
To detect multiple genes of smaller effects in predisposing individuals to Barrett’s esophagus.
Background
Barrett’s esophagus (BE) is a metaplastic premalignant disorder in which the normal stratified squamous epithelium of the lower esophagus is replaced by a columnar lined epithelium with intestinal differentiation. BE generally occurs in the context of chronic gastroesophageal reflux disease (GERD)Citation1 that can induce metaplastic change of the distal esophagus whereby the normal squamous epithelium is substituted by a columnar epithelium.Citation2
BE represents the primary risk factor for the development of esophageal adenocarcinoma (OAC) and is associated with an increased risk of cancer by about 0.4%–1% a year;Citation3,Citation4 the prevalence increases with age, comprising 1% of the population over 60 years of age. The disease is more common in men than in women (2:1),Citation5 it is rare in childhood, and it is estimated that the median age for the onset of the disease is approximately 40 years, although the median age at the time of diagnosis is approximately 60 years.Citation6 The risk of developing cancer is 30–125 times higher in BE patients as compared to the general population.Citation7
It has been clearly established that GERD is the main risk factor for the development of BE;Citation8 Caucasian males over the age of 50 with longstanding GERD are at higher risk of developing BE and OAC.Citation5,Citation9
The second most common condition associated with Barrett’s esophagus and OAC is central obesity.Citation10,Citation11 Nonetheless, since only a minority of subjects exposed to these environmental factors are found to have columnar metaplasia in the distal esophagus, BE is considered to be a complex disease in which the environment interacts with an individual’s genetic predisposition.Citation2
Over the last three decades, evidence has accumulated suggesting the presence of an inherited genetic component impacting on an individual’s predisposition to developing BE.Citation12 The age of occurrence of familial cases was found to be about 10 years younger with sporadic BE, and inheritance patterns of these families was consistent with an autosomal dominant disease with incomplete penetrance.
To date, relatively little is known about the specific genes that confer susceptibility to BE carcinogenesis. van Lieshout et alCitation13 studied the frequencies of polymorphic variants in glutathione S-transferase P1 (GSTP1) among 247 blood donors, 98 BE patients, and 34 subjects with OAC. They found an association between the GSTP1 polymorphic variant, GSTP1b, and an increased risk of both BE and OAC. In this study, subjects with BE and OAC were recruited through a single center rather than using a population-based approach, increasing the risk of selection bias. Abbas et alCitation14 carried out a case-control study of esophageal cancer in northwest France. They studied the frequency of the same GSTP1 variant but found no association between GSTP1 and risk of OAC in 27 patients. A number of factors, including inappropriate control group, lack of population-based DNA collections, and small study size, probably accounted for the discrepancies between the published studies.
In further studies, X-ray repair cross-complementing protein 1 (XRCC1) and the increased susceptibility for developing esophageal squamous cell carcinoma have been evaluated. However, the authors found that the XRCC1 399 Arg/Arg was found more frequently in patients with esophageal cancer who were alcohol drinkers; therefore, the results were not statistically significant. Citation15 In addition, the authors observed the genetic polymorphisms in XRCC1 associated with an increased risk of developing squamous cell carcinoma in a Chinese population.
Several other studies have reported an association between DNA repair genes and the pathogenesis of BE and OAC with a higher frequency of xeroderma pigmentosum complementation group C (XPC), poly AT insertion/deletion in OAC, a lower frequency of xeroderma pigmentosum complementary group D (XPD), and XRCC1 homozygous variants in BE, as compared with normal controls.Citation15–Citation17
Although the data noted in the literature are not always in accordance amongst several populations, the findings prompted us to carry out similar genetic studies in an Italian cohort of 74 patients with BE.
This preliminary study aimed to assess the association between genetic polymorphisms in DNA repair genes and genes with detoxifying enzymes on the development of BE, and subsequently with the increased risk for OAC. We studied six polymorphisms in four genes (XPC, XPD, XRCC1, GSTP) in 74 cases of patients with BE and 67 controls.
Materials and methods
Patients
The group of patients consisted of 74 individuals with a confirmed diagnosis of BE as per an identical protocol coming from six gastrointestinal units in Italy. Inclusion criteria were to be older than 18 years old and to have acquired a diagnosis of BE through endoscopy and histology. All of the cases belong to a wider epidemiological study aimed at identifying individual and environmental risk factors for BE.Citation18
A total of 67 healthy donor controls were recruited both from the gastrointestinal units described previously and from Niguarda Hospital (Milan, Italy). The selection criteria for control subjects included having no individual or family history of cancer. Controls were matched to patients for sex, age, and the same geographic origin. All control subjects were negative for BE.
The mean age was 63.08 years (SD = 13.74) for cases (53 male), and 58.57 years (SD = 11.16) for controls (43 males).
After giving written informed consent, all subjects provided peripheral blood samples.
Genotype analysis
DNA was extracted from whole blood using the QIAmp Blood Mini Kit (QIAGEN SRL, Milan, Italy).
Six different single nucleotide polymorphisms (SNPs) were investigated: XRCC1 arg194trp; XRCC1 arg399gln; XPC poly AT insertion/deletion indicated as pat; XPD arg156arg (allele C or A); GSTP1 Ile105Val; and Ala114Val. These latter SNPs can be combined to give different alleles with reduced enzymatic activity. Primers are available upon request.
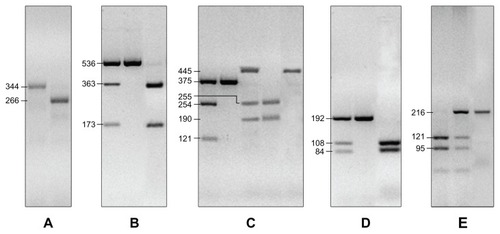
The XPC pat polymorphism was analyzed by polymerase chain reaction (PCR) and then resolved on a 2% agarose gel stained with Gel Red™ 10000X (Biotium, Hayward, CA). Homozygous pat −/− genotypes were represented by a 266 bp DNA band, whereas homozygous pat +/+ genotypes were represented by a 344 bp fragment; heterozygous +/− were represented by a 266 bp plus a 344 bp DNA fragment.
The polymorphisms in XPD and in XRCC1 were analyzed by PCR combined with restriction fragment length polymorphisms. The XPD PCR product was digested with Thermus filiformis, whereas restriction enzymes Proteus vulgaris 2 and methylation-specific PCR 1 were used to detect exon 6 and exon 10 polymorphisms in the XRCC1 gene, respectively (New England BioLabs GmbH, Frankfurt, Germany).
The GSTP1 polymorphisms in exon 5 and in exon 6 were assayed according to the methods and enzymes previously described.Citation19 Brief PCR amplifications were followed by enzymatic digestion with BsmAI and Acil, respectively (New England BioLabs GmbH) ().
Figure 1 Genotyping of XRCC1 arg194trp and arg399gln; XPC poly AT insertion/deletion indicated as pat; XPD arg156arg (allele C or A); GSTP1 Ile105Val and Ala114Val. (A) PCR products for XPC pat alleles: pat −/− generate the 266 bp fragment, pat +/+ generate the 344 bp fragment; heterozygous +/− were represented by a 266 bp plus a 344 bp DNA fragments. (B) The XPD arg156arg SNP is represented as nucleotide, which we identified in C/C alleles which gave 536 bp, C/A alleles with 536 bp, 363 bp and 173 bp fragments and A/A alleles with 363 bp and 173 bp fragments. (C) the XRCC1 arg194arg is indicated by a 375 bp fragment while the arg194trp is indicated by a 375 bp, 254 bp and 121 bp fragments. The XRCC1 arg399arg produced the 445 bp fragment, the arg399gln produced the 445 bp, 255 bp and 190 bp fragments, and gln399gln produced 255 bp and 190 bp fragments. (D) the SNPs in detoxifying gene GSTP1: ala114ala gave the 192 bp fragment, ala114val the 108 bp and 84 bp fragments; val114val the 192 bp, 108 bp and 84 bp fragments. (E) GSTP1 ile105ile was determined by the 216 bp fragment, the ile105val by the 121 bp and 95 bp fragments, the val105val by the 216 bp, 121 bp and 95 bp fragment.
Database
Each record corresponded to a known clinical condition (case) or to a sample population (control). These data comprised the variables that corresponded to the six SNPs, each of which could have three genotype classes: wild-type, heterozygous, and homozygous status. Each participant’s sex and smoking status information were included in the database.
Those genetic markers for which only one genotype was present both in cases and controls were removed from the database. There were 20 remaining variables in total in cases and controls.
Statistical analysis
The association of each tested variable (genetic polymorphisms, sex, and smoker status) with BE was tested using descriptive statistics including linear correlation and by using Artificial Neural Networks (ANNs). The models we used aimed to correctly classify the subjects according to one of two classes: Barrett’s esophagus patients (cases) or healthy subjects (controls).
No other specific genetic models potentially linked to the analyzed SNPs were evaluated. ANNs are able to build a model with a strong genetic basis by collecting all the information included within the SNP without any a priori definitions. The mathematical approach of ANNs consists of measuring the general dependence of random variables related to a group of subject without making any assumptions about the nature of their underlying relationships.Citation20,Citation21
Advanced intelligent systems, which are based on novel coupling of artificial neural networks and evolutionary algorithms, have also been applied in the present study. Supervised ANNs are networks which learn by examples and calculate an error function during the training phase and adjust the connection strengths in order to minimize the error function.Citation22 The learning constraint of the supervised ANNs makes their own output coincide with the predefined target. The general form of these ANNs is:
where y is the dependent variable; f is the mathematical function; x is the independent variables; and w the set of parameters that best approximates the function.
Data analysis was performed using a re-sampling system named TWIST, developed by the Semeion Research Center (Rome, Italy). The TWIST system consists of an ensemble of two previously described systems: Training and Testing (T and T) and Input Selection (IS).Citation23 The T and T system is a robust data re-sampling technique that is able to arrange the source sample into sub-samples that all possess a similar probability density function. In this way, the data is split into two or more sub-samples in order to train, test, and validate the ANN models more effectively. The IS system is an evolutionary wrapper system able to reduce the amount of data while conserving the largest amount of information available in the dataset. The combined action of these two systems allows us to solve two frequent problems in managing ANNs (ie, the optimal splitting of the data set in training and testing subsets containing a balanced distribution of outliers, and the optimal selection of variables) with the maximal amount of information relevant to the problem under investigation. Both systems are based on a genetic algorithm, the genetic doping algorithm developed at Semeion Research Center.Citation24 The TWIST system has been previously applied in different medical contexts;Citation25 additional data are given.
After this processing, the features that were most significant for the classification of patients into either the BE or control category were selected, and at the same time the training set and the testing set were created with a function of probability distribution similar to the one that provided the best results in the classification. A series of supervised multilayer perceptrons, with four hidden units, were then used for the classification task. The final ANNs, which were trained and tested on the new data set generated by the TWIST system, are “virgin,” and operate independently and blindly from each other and from the TWIST system.
Back-propagation ANNsCitation25 were then applied to the results obtained from the TWIST approach using a validation protocol. This is a procedure that is used to verify the model’s ability to generalize the results reached in the testing phase. Among the different protocols reported in literature, the selected model is the protocol with the greatest generalizability on data unknown to the model itself. The procedural steps in developing the validation protocol are: (1) Subdividing the dataset randomly into two sub-samples – the first called the Training Set, and the second called the Testing Set; (2) choosing a fixed ANN (and/or organism) which is trained on the Training Set. In this phase, the ANNs learn to associate the input variables with those that are identified as targets; (3) saving the weight matrix produced by the ANNs at the end of the training phase and freezing it with all of the parameters used for the training; (4) showing the Testing Set to the ANNs so that in each case the ANNs can express an evaluation based on the training just performed. This procedure takes place for each input vector, but every result (output vector) is not communicated to the ANNs. In this way, the ANNs are evaluated only in reference to the generalizability that it acquired during the training phase; and (5) constructing a new ANN with identical architecture to the previous ANN and repeating the procedure from point 1. This general training plan has been employed twice, obtaining two independent classification experiments: the first by training ANNs on subsamble A and testing them on subsample B, and the second by training ANNs on subsample B and testing them on subsample A.
Results
This study included a total of 74 patients with BE and 67 population controls, representing 52.5% and 47.5% of the overall sample, respectively.
We failed to find XPC pat +/− and XRCC1 trp194trp in both cases and controls, whereas GSTP1 val114val was found in among the control group only.
The distributions and confidence intervals (95%) of sex, smoking history, and polymorphisms among cases and controls are reported in ; there were no statistically significant differences among the two groups. The r-value from the linear correlation allowed us to discriminate between possible protective factors and possible risk factors as reported in ; however, the results are without statistical relevance. The results that were obtained provided a strong rationale by which to employ ANNs.
Table 1 Distributions of sex, smoking history, and polymorphisms among cases and controls
Table 2 r-value from linear correlation
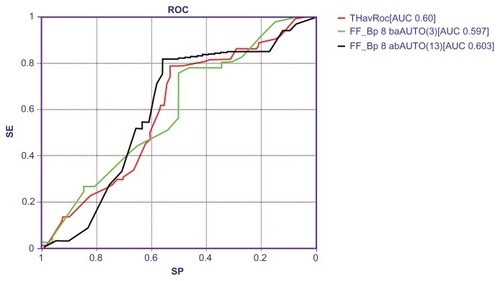
In using advanced intelligent systems like TWIST,Citation21 it was possible to reach the best predictive accuracy to discriminate between cases and controls. Indeed, the application of TWIST system allowed for the selection of a subgroup of nine variables, as reported in . This new data set has been analyzed with back-propagation ANNs employing a rigorous validation protocol. summarizes the results obtained with back-propagation ANNs applied 10 times on the final data set; a mean global accuracy of 60% was reached, and was as high as 65.88%.
Table 3 Variables selected by the TWIST system
Table 4 Classification performances of back-propagation neural networks on final data set
shows the area under the curve (AUC) of the receiver-operating characteristic (ROC), AUC of the two ANNs classifications, and the average ROC AUC.
Discussion
Barrett’s esophagus is a relatively common, benign, and asymptomatic disorder, the clinical importance of which relates to its role as a precursor lesion to esophageal adenocarcinoma; the condition appears to be a complex disease due not only to multiple genes/genetic variants, but also caused by environmental factors. Heavy, remote smoking, for example, has been associated with an increased risk of BE, suggesting a long latency period between exposure and development of the disease, even after smoking cessation.Citation26
Among the risk factors associated with the disease, case reports and pedigree studies suggest a heritable component, albeit with complex and variable expressions. A prominent feature of most cancers including Barrett’s adenocarcinoma is genetic instability, which is associated with the development and progression of the disease. Indeed, genetic instability has been shown to increase in patients with BE, involving less than 2% of the genome in the early stages to over 30% in later stages.Citation27
To safeguard the integrity of the genome, humans have developed a complex set of DNA repair systems. Defects in DNA repair have been demonstrated to be a critical mechanism in human carcinogenesis.Citation28 In addition, a reduced DNA repair capacity caused by genetic polymorphism is associated with an increased cancer risk.Citation29,Citation30 In this sense, numerous DNA polymorphisms have been identified in DNA repair genes, and many of them have been shown to contribute to genetic instability and error accumulation due to reduced protein activity.Citation31 These proteins are implicated in four major DNA repair pathways, including NER, BER, double-strand break repair, and mismatch repair.Citation32
Recently, the role of oxidative DNA damage, DNA repair, glutathione S-transferase Mu 1, superoxide dismutase 2, and 8-oxoguanine DNA glycosylase polymorphisms for individual susceptibility to BE have been investigated among 40 patients with BE. Even though the authors failed to find an association, the results of that study pointed to a role of oxidative DNA damage in BE.Citation33 To date, a large number of case-control studies to explore the association between DNA repair gene polymorphisms and the increasing risk of cancer have been performed. Several reports have shown an association between polymorphisms in the XPC and XPD genes and the increased risk of developing different types of cancer, with some XPD allelic variants related to an increased risk of lung cancer,Citation34 squamous cell carcinoma of the head and neck,Citation35 and breast cancer.Citation36
Several studies have also found associations between genetic polymorphisms in some BER genes, such as XRCC1, and an increased risk of cancer. BER genes play a key role in removing DNA damage from oxidation, deamination, and ring fragmentation,Citation37 and exposure to tobacco smoking induces oxidative damage by generating reactive oxygen species;Citation38 polymorphisms in BER genes have been shown to be associated with lung cancer.Citation31
Furthermore, given that exposure of esophageal epithelium to luminal toxic agents likely plays a crucial role, several studies have analyzed the association between polymorphisms in the detoxifying enzyme glutathione S-transferase and the risk of developing BE or OAC. GSTs comprise four main classes: A, M, P, and T, which are present in many species and tissues. Among them, the GSTP1 enzyme is the most important form found in the esophagus. Decreased GSTP1 enzyme activity has been detected in BE, suggesting that these alterations may contribute to an increased cancer risk in association with this disease.Citation39 Significantly lower GST enzyme activity was found more often in patients with BE and patients with OAC, indicating that these genetic changes may contribute to the development of both BE and OAC.Citation13
All the above data prompted us to investigate the XPC, XPD, XRCC1, and GSTP genetic polymorphisms in relation to Barrett’s esophagus in a cohort of Italian subjects. We report here results from an exploratory study involving 74 BE cases. As far as we know, this is the first Italian study with such a large cohort of subjects. All the samples were collected in gastrointestinal centers from the northern, central, and southern parts of Italy, and the clinical diagnoses have been unequivocally established.Citation17 Of the epidemiological data collected, only smoking status was analyzed together with the genetic data derived from analysis of the SNPs. We approached the dataset with classical statistical evaluation and with advanced intelligent systems. Indeed we believe that a non-conventional method, such as using advance intelligent systems, could successfully identify a genetic background (if present) that predisposes an individual to developing BE.
Results from this study failed to find an association among the tested SNPs and BE phenotype. We can hypothesize that the 74 cases tested here did not provide a large enough sample to find a statistically significant association given that the analysis may have been influenced by the number of tested variables and by the number of analyzed subjects. On the contrary, advanced intelligent systems such as TWIST system and back propagation are really able to handle the disease complexity, not treating the data with reductionist approaches unable to detect multiple genes of smaller effect in predisposing to the disease.
With such an approach, we were able to identify nine variables within the genes involved in the NER and BER DNA repair pathways and in a gene coding for detoxifying enzymes GSTP1. Interestingly, the XPC pat variable lies within chromosome 3p, a fragile region recently found to be involved in the early stages of BE.Citation40 However, back-propagation analysis on the variables selected by the TWIST system was not able to exceed a mean sensitivity of 78.71% with a mean specificity of 53.05%.
We can postulate that the genetic variables analyzed here do not represent the factors that make an individual susceptible to developing BE; however, our results suggest a strong positive correlation between genetic background and BE does not exist. Recently, residual embryonic cells have been proposed as a precursor of Barrett’s-like metaplasia, suggesting that this precancerous lesion originates not from genetic alterations, but from competitive interactions between cell lineages driven by opportunity.Citation41
Acknowledgments
We thank the research centers for patient enrollment, and we are grateful to all of the study participants for their cooperation. This work was supported by Bracco SPA. The Barrett Italian Study Group is: Antonella De Ceglie (Unit of Digestive Endoscopy, Cancer Institute Giovanni Paolo II, Bari), Rosa Filiberti and Vincenzo Fontana (Epidemiology, Biostatistic and Clinical Trials, National Institute for Cancer Research, Genova), Massimo Conio and Sabrina Blanchi (Department of Gastroenterology, General Hospital, Sanremo, Imperia, Italy), Enzo Grossi (Medical Department, Bracco Spa, Milan, Italy), Teresa Lacchin (Department of Gastroenterology and Digestive Endoscopy, IRCCS, Aviano, Pordenone, Italy), Marina De Matthaeis (Department of Gastroenterology and Digestive Endoscopy, Regina Margherita Hospital, Rome), Orazio Ignomirelli (Endoscopy Unit, IRCCS CROB, Rionero in Vulture, Potenza), Roberta Cappiello (Unit of Gastroenterology, S Maria degli Angeli Hospital, Pordenone), Domenico Della Casa (Unit of Digestive Endoscopy, University of Brescia, Brescia), Monica Foti (Department of Gastroenterology, Mauriziano Hospital, Torino), Francesco Laterza (Department of Internal Medicine and Gastroenterology, G D’Annunzio University & Foundation, Chieti), Riccardo Rosati (Invasive Surgery, IRCCS Clinical Institute Humanitas, Rozzano, Milano), Vito Annese (Unit of Gastroenterology and Digestive Endoscopy, IRCCS, Casa Sollievo della Sofferenza, S Giovanni Rotondo, Foggia), and Gaetano Iaquinto (Unit of Gastroenterology and Digestive Endoscopy, SG Moscati Hospital, Avellino).
Disclosure
The authors report no conflicts of interest in this work.
References
- BarberaMFitzgeraldRCCellular mechanisms of Barrett’s esophagus developmentSurg Oncol Clin N Am200918339341019500732
- di PietroMFitzgeraldRCBarrett’s oesophagus: an ideal model to study cancer geneticsHum Genet2009126223324619365640
- ShaheenNJCrosbyMABozymskiEMSandlerRSIs there publication bias in the reporting of cancer risk in Barrett’s esophagus?Gastroenterology2000119233333810930368
- YousefFCardwellCCantwellMMGalwayKJohnstonBTMurrayLThe incidence of esophageal cancer and high-grade dysplasia in Barrett’s esophagus: a systematic review and meta-analysisAm J Epidemiol2008168323724918550563
- WongNAWildingJBartlettSCDX1 is an important molecular mediator of Barrett’s metaplasiaProc Natl Acad Sci U S A2005102217565757015894614
- PondugulaKWaniSSharmaPBarrett’s esophagus and esophageal adenocarcinoma in adults: long-term GERD or something else?Curr Gastroenterol Rep20079646847418377797
- KruijshaarMESiersemaPDJanssensACKerkhofMSteyerbergEWEssink-BotMLCYBAR Study GroupPatients with Barrett’s esophagus perceive their risk of developing esophageal adenocarcinoma as lowGastrointest Endosc2007651263017185076
- VaeziMFRichterJERole of acid and duodenogastroesophageal reflux in gastroesophageal reflux diseaseGastroenterology19961115119211998898632
- WongAFitzgeraldRCEpidemiologic risk factors for Barrett’s esophagus and associated adenocarcinomaClin Gastroenterol Hepatol20053111015645398
- CorleyDAKuboALevinTRAbdominal obesity and body mass index as risk factors for Barrett’s esophagusGastroenterology20071331344117631128
- EdelsteinZRFarrowDCBronnerMPRosenSNVaughanTLCentral adiposity and risk of Barrett’s esophagusGastroenterology20071332403411 Epub May 21, 200717681161
- RobertsonEVJankowkiJAGenetics of gastroesophageal cancer: paradigms, paradoxes, and prognostic utilityAm J Gastroenterol20081032443449 Epub October 9, 200717925001
- van LieshoutEMRoelofsHMDekkerSPolymorphic expression of the glutathione S-transferase P1 gene and its susceptibility to Barrett’s esophagus and esophageal carcinomaCancer Res19995935865899973204
- AbbasADelvinquiereKLechevrelMGSTM1, GSTT1, GSTP1 and CYP1A1 genetic polymorphisms and susceptibility to esophageal cancer in a French population: different pattern of squamous cell carcinoma and adenocarcinomaWorld J Gastroenterol200410233389339315526353
- HaoBWangHZhouKIdentification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinomaCancer Res200464124378438415205355
- CassonAGZhengZEvansSCVeugelersPJPorterGAGuernseyDLPolymorphisms in DNA repair genes in the molecular pathogenesis of esophageal (Barrett) adenocarcinomaCarcinogenesis20052691536154115878910
- ShenHXuYQianYPolymorphisms of the DNA repair gene XRCC1 and risk of gastric cancer in a Chinese populationInt J Cancer20008860160611058877
- GrossiEFilibertiRBlanchiSW1995 artificial neural networks allow good discrimination of Barrett’s esophagus from GERD on the basis of biographic, clinical history, symptoms and lifestyle habitsGastroenterology20091365A-769
- LeeYLLinYCLeeYCWangJYHsiueTRGuoYLGlutathione S-transferase P1 gene polymorphism and air pollution as interactive risk factors for childhood asthmaClin Exp Allergy200434111707171315544594
- GrossiEArtificial Adaptive Systems and predictive medicine: a revolutionary paradigm shiftImmun Ageing20107Suppl 1S321172062
- GrossiEBuscemaMIntroduction to artificial neural networksEur J Gastroenterol Hepatol200719121046105417998827
- RumelhartDEMcClellandJLParallel Distributed Processing: Explorations in the Microstructure of Cognition1Cambridge, MAFoundations, MIT Press1986
- BuscemaMGrossiEIntraligiMGarbagnaNAndriulliABredaMAn optimized experimental protocol based on neuro-evolutionary algorithms application to the classification of dyspeptic patients and to the prediction of the effectiveness of their treatmentArtif Intell Med200534327930516023564
- BuscemaMGenetic Doping Algorihm (GenD): theory and applicationsExpert Systems20042126379
- BuscemaMCapriottiMBergamiFBabiloniCRossiniPGrossiEThe implicit function as squashing time model: a novel parallel nonlinear EEG analysis technique distinguishing mild cognitive impairment and Alzheimer’s disease subjects with high degree of accuracyComput Intell Neurosci2007 Article ID 3502115
- JacobsonBCGiovannucciELFuchsCSSmoking and Barrett’s esophagus in women who undergo upper endoscopyDig Dis Sci20115661707171721448698
- LiXGalipeauPCSanchezCASingle nucleotide polymorphism-based genome-wide chromosome copy change, loss of heterozygosity, and aneuploidy in Barrett’s esophagus neoplastic progressionCancer Prev Res (Phila)20081641342319138988
- HoeijmakersJHGenome maintenance mechanisms for preventing cancerNature2001411683536637411357144
- BenhamouSSarasinAVariability in nucleotide excision repair and cancer risk: a reviewMutat Res20004622–314915810767626
- BerwickMVineisPRESPONSE re: markers of DNA repair and susceptibility to cancer in humans: an epidemiologic reviewJ Natl Cancer Inst2000921887489710841823
- López-CimaMFGonzález-ArriagaPGarcia-CastroLPolymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern SpainBMC Cancer2007716217705814
- WoodRDMitchellMSgourosJLindahlTHuman DNA repair genesScience200129155071284128911181991
- KadiogluESardasSErgunMUnalSKarakayaAEThe role of oxidative DNA damage, DNA repair, GSTM1, SOD2 and OGG1 polymorphisms in individual susceptibility to Barrett’s esophagusToxicol Ind Health2010262677920056743
- ZienolddinySCampaDLindHPolymorphisms of DNA repair genes and risk of non-small cell lung cancerCarcinogenesis200627356056716195237
- SturgisEMZhengRLiLXPD/ERCC2 polymorphisms and risk of head and neck cancer: a case-control analysisCarcinogenesis200021122219222311133811
- TerryMBGammonMDZhangFFPolymorphism in the DNA repair gene XPD, polycyclic aromatic hydrocarbon-DNA adducts, cigarette smoking, and breast cancer riskCancer Epidemiol Biomarkers Prev200413122053205815598760
- FrosinaGCommentary: DNA base excision repair defects in human pathologiesFree Radic Res200438101037105415512792
- WilsonDM3rdSofinowskiTMMcNeillDRRepair mechanisms for oxidative DNA damageFront Biosci20038d963d98112700077
- BrabenderJLordRVWickramasingheKGlutathione S-transferase-pi expression is downregulated in patients with Barrett’s esophagus and esophageal adenocarcinomaJ Gastrointest Surg20026335936712022988
- LaiLAKostadinovRBarrettMTDeletion at fragile sites is a common and early event in Barrett’s esophagusMol Cancer Res2010881084109420647332
- WangXOuyangHYamamotoYResidual embryonic cells as precursors of a Barrett’s-like metaplasiaCell201114571023103521703447