Abstract
The protein encoded by the TP53 gene is one of the most important suppressors of tumor formation, which is also frequently inactivated in gastrointestinal cancer. MicroRNAs (miRNAs) are small noncoding RNAs that inhibit translation and/or promote degradation of their target messenger RNAs. In recent years, several miRNAs have been identified as mediators and regulators of p53’s tumor suppressing functions. p53 induces expression and/or maturation of several miRNAs, which leads to the repression of critical effector proteins. Furthermore, certain miRNAs regulate the expression and activity of p53 through direct repression of p53 or its regulators. Experimental findings indicate that miRNAs are important components of the p53 network. In addition, the frequent genetic and epigenetic alterations of p53-regulated miRNAs in tumors indicate that they play an important role in cancer initiation and/or progression. Therefore, p53-regulated miRNAs may represent attractive diagnostic and/or prognostic biomarkers. Moreover, restoration of p53-induced miRNAs results in suppression of tumor growth and metastasis in mouse models of cancer. Thus, miRNA-based therapeutics may represent a feasible strategy for future cancer treatment. Here we summarize the current published state-of-the-art on the role of the p53-miRNA connection in gastrointestinal cancer.
Keywords:
Introduction
Gastrointestinal (GI) cancers represent malignant tumors of the GI tract and accessory organs of digestion. They include carcinomas arising in the oral cavity, esophagus, stomach, liver, gallbladder, pancreas, small intestine, large intestine, rectum, and anus. GI cancer represents about 30% of all tumor incidences and is responsible for approximately 40% of tumor-related mortality worldwide ().Citation1 Tumors of the GI tract harbor mutations in the p53 tumor suppressor gene (TP53) with a prevalence ranging from 31% to 45% ().Citation2 The p53 protein functions as a transcription factor that mediates the response to many cellular stresses, most prominently the DNA damage response. p53 suppresses a variety of malignant processes, thereby representing one of the most important cancer suppressing proteins.Citation3 p53 protects against cancer by inducing cellular processes such as apoptosis or cell cycle arrest, thereby preventing the propagation of damaged cells that potentially could give rise to tumors.Citation4,Citation5 Moreover, p53 inhibits epithelial to mesenchymal transition (EMT), stemness, and metabolic adaptations, which are typically found in tumors.Citation6 In addition, p53 promotes DNA repair, antioxidant defense, and differentiation. On the molecular level, p53 exerts its tumor suppressive functions by regulating the expression of numerous target genes, mainly by direct binding to specific DNA motifs located in target gene promoters.Citation7 Besides p53-regulated protein expression, p53-induced microRNAs (miRNAs) have emerged as important effectors of p53.Citation8,Citation9 The generation of mature miRNAs is a multistage process (see ) starting with the transcription of miRNA encoding genes to yield the primary miRNA (pri-miRNA).Citation10 Next, the pri-miRNA is cleaved by the RNAse III enzyme Drosha, resulting in a ~70 nucleotide stem-loop-structured miRNA precursor molecule (pre-miRNA).Citation11 The pre-miRNA is transported to the cytoplasm by Exportin 5, where it is cleaved further by the RNAse Dicer. The resulting 20 to 25 bp mature miRNA is incorporated into the miRNA-induced silencing complex (miRISC), which mediates miRNA-induced silencing of target messenger RNAs (mRNAs).Citation12 miRNAs bind to 3′-untranslated regions (3′-UTR) of mRNAs via their seed sequences, which are conserved seven nucleotide regions in their 5′ region. The association of the miRISC with seed-matching sequences in target mRNAs results in the inhibition of translation and degradation of the target mRNAs.Citation10 It has been estimated that >60% of human protein coding genes are subject to regulation by miRNAs.Citation13 Not surprisingly, miRNA-mediated regulation has been implicated in almost all physiological and pathophysiological processes.Citation10 Interestingly, several miRNAs may also be of use for diagnostic, prognostic, and therapeutic applications in GI-cancers.Citation14–Citation18 Extracellular miRNAs have been detected in blood serum. These miRNAs are either secreted by living cells via exosomes or microvesicles, or they originate from dying cells.Citation19 Interestingly, these circulating miRNAs are extremely stable, both in blood and after isolation. Numerous studies have shown their potential usefulness as noninvasive diagnostic and prognostic markers in GI cancers.Citation20 Seven years ago it was shown that p53 also regulates the expression of miRNA-encoding genes.Citation8 The p53-regulated miRNAs have been implicated in the control of various cancer-related processes, such as proliferation, apoptosis, EMT, migration, invasion, and metastasis. Therefore, they may represent important mediators of the tumor suppressive function of p53. In addition, a number of miRNAs can regulate expression and activity of the p53 protein, either negatively through direct repression of p53 expression, or positively through the repression of negative regulators of p53. In this review we summarize the current knowledge about the p53/miRNA network and its role in GI cancers.
Figure 1 Incidence (A) and mortality (B) of indicated gastrointestinal (GI) cancers worldwide. (C) Prevalence of mutations in the TP53 gene in the indicated GI cancers.
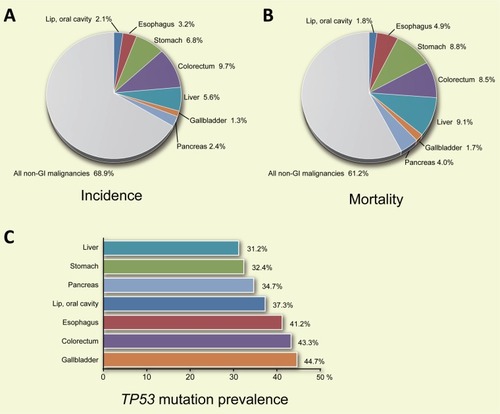
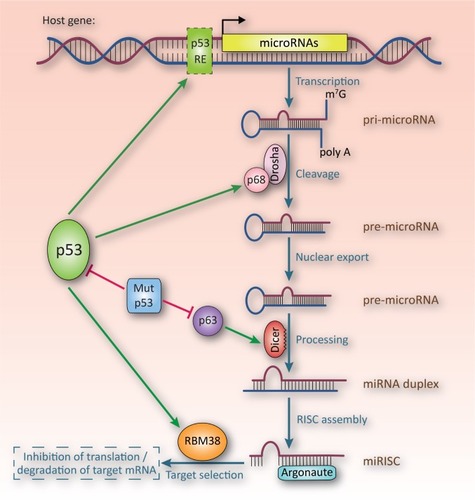
Figure 2 Effects of p53 on miRNA biogenesis.
Abbreviations: mRNA, messenger RNA; miRNA, microRNA; pre-microRNA, miRNA precursor molecule; pri-microRNA, primary miRNA; miRISC, miRNA-induced silencing complex; mut, mutant; RE, response element.
p53 regulated miRNAs
In 2007, we and other groups identified several miRNAs as direct transcriptional targets of p53.Citation21–Citation27 Since then, many of these miRNAs have been validated as important mediators of p53 functions ( and ).Citation9 p53 regulates the expression of its target miRNAs either on the transcriptional level by direct binding to the promoters of the corresponding genes, or by regulating miRNA processing (). It was shown that p53 interacts with the DEAD-box RNA helicase p68 (also known as DDX5) and enhances its interaction with the Drosha complex. As a result, p53 promotes the processing of specific pri-miRNAs to pre-miRNAs, leading to elevated levels of the respective miRNAs.Citation28 Another link between p53 and miRNA-processing has been observed in conditional Dicer knockout mice.Citation29 Dicer deficiency, and therefore incomplete miRNA maturation, induces p53, which leads to reduced proliferation and premature senescence. Therefore, p53 may operate as a checkpoint to monitor proper miRNA processing. Moreover, expression of Dicer1 is regulated by the p53 family member p63, which can be inhibited by association with mutant p53.Citation30 Finally, p53 also affects miRNA target gene selection by regulating mRNA-binding proteins, such as RNA-binding-motif protein 38, which competes with miR-NAs for binding to 3′-UTRs of mRNAs.Citation31 miRNAs transcriptionally induced by p53 include the miR-34,Citation21–Citation26 miR-200,Citation32 miR-15a/16-1,Citation33 and miR-192/194/215Citation34 clusters, as well as miR-145Citation35 and miR-107.Citation36 Yet some of these miRNAs, such as miR-16-1, miR-145, and miR-199a-3p, are also regulated on the post-transcriptional level by p53.Citation28,Citation37 The expression of the majority of these miRNAs is frequently altered in GI tumors and has been associated with clinical and pathological parameters of various types of GI cancer ( and ).
Table 1 Summary of changes in expression of p53-pathway-related miRNAs in GI cancers
Table 2 Compilation of p53-regulated miRNAs and their alterations in GI cancers
Table 3 Compilation of miRNAs that directly target p53 and their alterations in GI cancers
The miR-34 family
The miR-34 family includes three members – miR-34a, miR-34b, and miR-34c – which show a marked induction by p53.Citation8 MiR-34a is encoded by its own host gene, whereas miR-34b and miR-34c share a common precursor. Both miR-34 genes contain several p53-responsive elements, which are occupied by p53 and mediate activation of miR-34a/b/c after DNA damage.Citation25,Citation26 Expression of miR-34a/b/c is frequently downregulated in colorectal,Citation38 esophageal,Citation39 gastric,Citation40 and hepatocellular cancers (HCCs).Citation41 Consistently, all members of the miR-34 family were shown to suppress tumor growth and metastasis by inhibiting processes that promote cancer, including cell cycle progression, EMT, metastasis, and stemness and by promoting tumor suppressive processes, such as apoptosis and senescence.Citation42 MiR-34s regulate these processes through suppressing the expression of their target mRNAs, such as SNAIL, c-Myc, Bcl2, c-Met, and Axl.Citation43 The miR-34/p53 axis and its targets are often connected through positive or negative feedback loops that either reinforce the miR-34/p53 signaling or suppress it. For example, a positive feedback loop connects miR-34a and p53 via MDM4 (). MDM4 and its human counterpart HDM4 bind to p53 and inhibit its transcriptional activity. At the same time MDM4/HDM4 are targets of miR-34a.Citation44,Citation45 Therefore, the repression of MDM4/HDM4 by miR-34a leads to stabilization of p53 and enhanced expression of miR-34a. Recently, it was shown that in addition to full-length HDM4 that is targeted by miR-34a, a short isoform of HDM4 also exists, which lacks seed-matching sites for miR-34a, thereby evading suppression by miR-34a.Citation45 Consistently, this short HDM4 isoform was highly expressed in tumors, where it presumably inhibits the miR-34a/p53 axis.
Figure 3 The role of p53/miRNA axis in (A) p53 autoregulation, (B) cancer cell metabolism, (C) invasion, and metastasis, as a result of the regulation of EMT/MET (D) cancer-associated inflammatory signaling.
Abbreviations: EMT, epithelial–mesenchymal transition; IL-6, interleukin 6; IL-6R, interleukin 6 receptor; MET, mesenchymal–epithelial transition; miRNA, microRNA; NAD+, nicotinamide adenine dinucleotide; NAMPT, nicotinamide phosphoribosyltransferase.
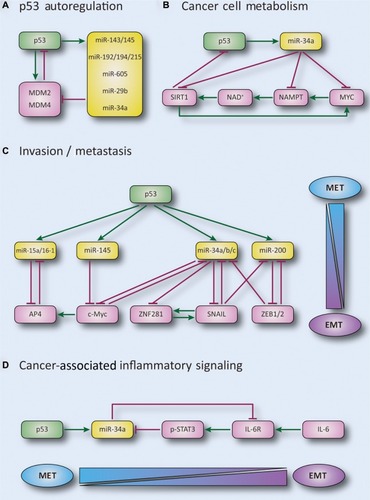
Furthermore, SIRT1 was shown to mediate activation of p53 by miR-34a.Citation46 SIRT1 is an nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase, which represses p53 activity by deacetylation of p53 protein. Yamakuchi et al showed that SIRT1 is a miR-34 target and that miR-34 induces the activity of p53 by repressing SIRT1 in colorectal cancer (CRC) cell lines (). Moreover, miR-34 not only represses the expression of SIRT1, but also suppresses SIRT1 activity by downregulating nicotinamide phospho-ribosyltransferase (NAMPT), the rate-limiting enzyme of NAD+ biosynthesis.Citation47 Furthermore, SIRT1 and MYC regulate each other via a positive feedback loop.Citation48,Citation49 By repressing both SIRT1 and MYC, miR-34a may therefore efficiently suppress this circuitry.
Another double-negative feedback loop involving miR-34a was discovered by Siemens et al,Citation50 who demonstrated that miR-34a directly targets and suppresses the EMT-inducing transcription factor (EMT-TF) SNAIL, whereas SNAIL represses the miR-34a and miR-34b/c genes by directly binding to their promoters in CRC cell lines (). By utilizing HCC and CRC cells, Kim et al showed that p53 regulates EMT by inducing members of the miR-200 family,Citation32 which also represent EMT-regulating miRNAs that suppress EMT by a similar double-negative feedback loop involving the EMT-TFs ZEB1 and ZEB2.Citation51,Citation52 Thus, p53 is a key regulator of cellular plasticity by controlling EMT and its counterpart mesenchymal–epithelial transition (MET) through inducing the miRNAs of the miR-34 and miR-200 family (). These miRNAs form two double-negative feedback loops with their targets SNAIL, ZEB1, and ZEB2 that act as bimodal switches to stabilize either the epithelial or the mesenchymal state. Moreover, ZEB1 was also shown to repress miR-34a expression by binding to the same E-boxes in the miR-34 promoters as SNAIL, thereby further interconnecting the miR-34/SNAIL and miR-200/ZEB loops.Citation50,Citation53 In addition, we recently showed that the zinc-finger 281 (ZNF281) protein is an important miR-34 target with respect to EMT.Citation54 We found that the expression of ZNF281 is controlled by miR-34 and SNAIL in a coherent feed-forward-loop, whereby SNAIL and ZNF281 induce each other, whereas miR-34 can directly repress both SNAIL and ZNF281 (). Accordingly, ectopic SNAIL induced EMT by directly activating ZNF281 and concomitantly repressing miR-34a expression, which leads to a further increase in ZNF281 levels. Notably, the induction of ZNF281 by SNAIL was required for SNAIL-induced EMT.
Recently, we demonstrated that inflammation can suppress the expression of miR-34a.Citation55 We showed that exposure to the proinflammatory cytokine interleukin-6 (IL-6) results in repression of miR-34a via direct binding of the IL-6 effector STAT3 to the first intron of the miR-34a gene. Furthermore, we identified the IL-6 receptor (IL-6R) as a direct target of miR-34a. Further functional analysis revealed the existence of an IL-6R/STAT3/miR-34a feedback loop (). The activation of this loop was required for EMT, invasion, and metastasis of CRC cell lines and was associated with nodal and distant metastasis in CRC patients. Moreover, in miR-34a-deficient mice, colitis-associated intestinal tumors displayed activation of the loop and, in contrast to tumors in wild-type mice, progressed to invasive carcinomas. Our findings suggest that the activation of the IL-6R/STAT3/miR-34a loop by IL-6 drives cancer cells toward a mesenchymal and invasive phenotype, whereas suppression of the loop by p53 shifts cancer cells toward an epithelial state and prevents EMT and invasion.Citation55
Reintroduction of miR-34 into tumors, which lost miR-34 expression, may represent an attractive alternative for cancer treatment. The most common approach for miRNA delivery relies on lipid-based nanoparticles, which contain vectors expressing miRNAs, or ∼19–23-nt double-stranded mimics of mature miRNAs. These can be administered systemically by intravenous injection or locally into tumors. Several studies have shown that systemic miR-34a delivery suppresses tumor growth in vivo. Using xenograft or genetically engineered mouse models of melanoma, lymphoma, multiple myeloma, breast, prostate, pancreatic, and non-small-cell lung cancer, the authors observed an inhibition of tumor growth by 20% to 83% after reintroduc tion of miR-34a.Citation56 Importantly, no severe toxicity caused by systemic miR-34a delivery has been observed in mice.Citation57,Citation58 Likewise, no unwanted immune response has been detected, based on serum cytokine levels in immune-competent mice.Citation59 Recently, the company Mirna Therapeutics has initiated a clinical Phase I trial of nanoparticle-based delivery of miR-34a in patients with non-resectable primary liver cancer or metastatic cancer with liver involvement.Citation60 Therefore, miR-34a may be one of the first miRNA mimics to reach the clinic. Conventional anticancer therapies, such as chemotherapy and treatment with radiation, induce miR-34 expression in human cancer cells with wild-type p53.Citation27 However, since the majority of human tumors lack normal p53 function, replacement of miR-34 may enhance the efficacy of standard cancer therapies. Indeed, in prostate, colorectal, and bladder cancer cells, reintroduction of miR-34a precursors enhanced the sensitivity toward camptothecin, paclitaxel, 5-fluorouracil, and cisplatin.Citation61–Citation65 Furthermore, lentiviral transduction of miR-34a sensitized gastric and pancreatic cancer cells to radiation and to the chemotherapeutic drugs docetaxel, gemcitabine, cisplatin, and doxorubicin.Citation63,Citation64 Moreover, we recently showed that c-Kit is an important miR-34a target that mediates, at least in part, chemosensitization by miR-34a in CRC cell lines.Citation65 Thus, these results suggest that combined treatment with miR-34 mimics may enhance the beneficial effects of conventional cancer therapies.
Downregulation of miR-34 expression in tumors has been frequently attributed to methylation of the CpG islands present in promoters of miR-34a and miR-34b/c.Citation66–Citation69 CpG methylation is causally involved in repression of miR-34-a/b/c, since treatment of CRC cell lines with the demethylating agent 5-aza-2′-deoxycytidine leads to re-expression of miR-34a/b/c.Citation66,Citation68,Citation69 Moreover, a significant inverse correlation between miR-34a methylation and expression has been observed in colon tumors.Citation70,Citation71 Therefore, miRNA cancer treatment strategies may rely not only on delivery of synthetic miRNA mimics corresponding to miR-34a/b/c into tumors, but also on re-expression of these miRNAs using demethylating agents. Indeed, treatment with BioResponse 3,3′-Diindolylmethane, an experimental anti-androgen prostate cancer drug, resulted in demethylation and re-expression of miR-34a in prostate cancer cells.Citation72 In a Phase II clinical trial, treatment of prostate cancer patients with BioResponse 3,3′-Diindolylmethane prior to radical prostatectomy led to the re-expression of miR-34a as well as repression and nuclear exclusion of its target, the androgen receptor.Citation72,Citation73 Several reports showed that miR-34 methylation may also have prognostic value. In our study, miR-34a methylation in primary CRC was significantly associated with increased formation of lymph node and liver metastases.Citation70 Recently, Wu et al analyzed miR-34a/b/c methylation in stool samples of 82 CRC patients and 40 controls.Citation74 They demonstrated that detection of miR-34a and miR-34b/c methylation could identify CRC with a remarkable sensitivity of 76.8% or 95% and a specificity of 93.6% or 100%, respectively. Therefore, the detection of miR-34 miRNAs and CpG methylation of miR-34 promoter regions in body fluids or stool represent potential biomarkers which may be utilized for noninvasive screening and diagnosis of cancer in the future.
miR-15a and miR-16-1
Mir-15a and miR-16-1 were the first miRNAs genetically linked to cancer: In 2002, Calin et al showed that miR-15a and miR-16-1, which are encoded within an intron of the DLEU2 gene, are frequently deleted and/or downregulated in chronic lymphocytic leukemia.Citation75 Notably, a knockout of DLEU2 or of the miR-15a/16-1-bearing intron in mice confirmed that loss of miR-15a/16-1 causes chronic lymphocytic leukemia.Citation76 The expression of miR-15a and miR-16-1 is induced by p53 via transcriptionalCitation33 and post-transcriptional mechanisms.Citation28 Several studies implicated the downregulation or loss of miR-15a and miR-16-1 expression in GI cancers. For example, the expression of miR-16-1 was significantly lower in primary CRC when compared to the corresponding normal colonic mucosa.Citation77 Moreover, decreased miR-16-1 expression was associated with lymph node metastasis and recurrence of colorectal tumors.Citation77 Furthermore, ectopic expression of miR-15a and miR-16-1 inhibited the proliferation of pancreatic and colorectal cancer cellsCitation78,Citation79 and led to a significant inhibition of subcutaneous growth of CRC cell lines in immune-compromised mice.Citation80 Interestingly, hepatitis B virus X protein, which is involved in the initiation and progression of HCC downregulates miR-15a/16 expression, suggesting that reintroduction of these miRNAs may be an effective treatment of hepatitis-B-virus-related chronic liver diseases.Citation81 MiR-15a and miR-16-1 act tumor suppressive, at least in part, by promoting apoptosis and cell cycle inhibition via targeting the anti-apoptotic protein Bcl2Citation82 and cell cycle regulators, including CDK6 and cyclin D1, respectively.Citation83,Citation84 Recently, we demonstrated that miR-15a/16-1 also inhibit EMT, invasion, and metastasis of CRC cells by directly targeting the EMT-TF AP4.Citation85 Interestingly, AP4 itself is a repressor of the DLEU2 gene. We showed that miR-15a/16-1 and the EMT-inducing factor AP4Citation86 form a double-negative feedback loop that stabilizes low expression of miR-15a/16-1 and elevated expression of AP4 in invasive CRC cells and tumors, thereby ultimately promoting CRC metastasis ().Citation85
miR-145
p53 controls the expression of miR-145 by two mechanisms: first, p53 directly induces the transcription of the miR-145 gene,Citation35 and second, p53 enhances miR-145 maturation via modulation of Drosha-mediated miRNA processing.Citation28,Citation87 In line with a regulation by p53, expression of miR-145 is significantly lower in various tumors that harbor p53 mutations, including esophageal, gastric, pancreatic, colorectal, and bladder cancers.Citation88–Citation92 Accordingly, ectopic miR-145 suppresses migration, invasion, and metastasis of gastric and colorectal cancer cells.Citation89,Citation93 Moreover, therapeutic polyethylenimine-mediated reintroduction of miR-145 reduces proliferation and increases apoptosis of CRC cells in xenograft mouse models.Citation94 The tumor suppressing properties of miR-145 can be partially attributed to the repression of MYC, which represents a direct target of miR-145.Citation35 Similar to miR-34, miR-200, and miR-15a/16-1, miR-145 has also been shown to represent a mediator of p53-induced MET, the reversion of EMT ().Citation95 Another important oncogenic target of miR-145 is KRAS.Citation96 Interestingly, activated KRAS also represses miR-145 via RREB1, thereby forming a feed-forward loop that potentiates RAS signaling. Accordingly, loss of miR-145 is frequently observed in KRAS mutant pancreatic cancers, and restoration of these miRNAs inhibits tumorigenesis.Citation96 Xu et al showed that miR-145 negatively regulates the pluripotency factors OCT4, SOX2, and KLF4, and thereby represses self-renewal and induces differentiation of human embryonic stem cells.Citation97 Moreover, the same group reported that the miR-145 promoter is bound and repressed by OCT4, thereby forming a negative feedback loop.Citation97 Loss of p53 leads to increased generation of induced pluripotent stem cells and expansion of cancer stem cells.Citation98 This effect might at least in part be due to the lack of p53-induced miR-145 expression and consequent upregulation of OCT4. Finally, like miR-34a targets MDM4, miR-145 directly targets and represses the p53 inhibitor MDM2.Citation99 The result is another positive feed-forward loop that leads to stabilization of p53 and elevated expression of p53-induced miRNAs ().
The miR-192/miR-194/miR-215 family
MiR-192, miR-194, and miR-215 are encoded by two clusters located at two different sites: The miR-194-1/miR-215 cluster on chromosome 1 (1q41) and the miR-192/miR-194-2 cluster on chromosome 11 (11q13.1). Both clusters are directly induced by p53.Citation34,Citation100 Interestingly, miR-194-1 and miR-194-2 have the same mature sequence, although they are derived from two different precursors on two chromosomal locations. MiR-192 and miR-215 have the same seed sequence, whereas the seed sequence of miR-194 differs. All three miRNAs display decreased expression in colorectal tumors.Citation34 Furthermore, low expression of miR-194 and miR-215 significantly correlates with a high probability of relapse and shorter survival in colorectal patients.Citation101 MiR-192, miR-194, and miR-215 regulate cell cycle progression and proliferation via the repression of functionally important targets, such as CDC7, MAD2L1, and CUL5.Citation102 Moreover, miR-192 suppresses liver metastasis of CRC cells by targeting BCL2, ZEB2, and VEGF-A,Citation103 while miR-194 inhibits EMT in endometrial cells by targeting BMI-1.Citation104 Importantly, similar to other p53-induced miRNAs, miR-192, miR-194, and miR-215 also directly target MDM2 and therefore interfere with the autoregulatory MDM2/p53 loop ().Citation100
The miR-200 family
The two genes that give rise to the miR-200c/141 and the 200a/200b/429 miRNAs represent direct p53 target genes.Citation32 The members of the miR-200 family are tumor suppressing miRNAs and several studies showed that they play a crucial role in regulating the balance between EMT and MET by forming a double-negative feedback loop with their targets, the EMT-inducers ZEB1 and ZEB2 ().Citation52,Citation105 Moreover, miR-200c also suppresses stemness by targeting the stem cell factors KLF4, SOX2, and the polycomb repressor BMI-1.Citation106,Citation107 Several studies reported that elevated levels of cell-free, circulating miR-200c and miR-200a in the blood of colorectal, gastric, and esophageal cancer patients are associated with increased tumor stage, presence of metastases, and poor survival.Citation108–Citation112 At first sight, this data seems contradictory, since functional studies showed that miR-200s repress EMT, invasion, and metastasis. Yet, recent studies showed that during the formation of metastases, cancer cells undergo MET and re-express EMT-suppressing genes and miRNAs.Citation113 Therefore, elevated levels of EMT-suppressing circulating miRNAs, such as miR-200c, might originate from metastases and indicate metastatic dissemination.
miR-107
The miRNA miR-107 is encoded by an intron of the p53-induced pantothenate kinase 1 (PANK1) gene.Citation36,Citation114 Several studies showed that ectopic expression of miR-107 enhances EMT, migration, and promotes metastatic dissemination, whereas the loss of miR-107 represses migration and metastasis of colorectal, breast, and gastric cancer cells.Citation115–Citation118 In line with these observations, expression of miR-107 is higher in gastric tumors compared with adjacent normal tissue.Citation119 Moreover, high expression of miR-107 correlates with lymph node and distant metastasis as well as poor survival of colorectal, breast, and gastric cancer patients.Citation115–Citation120 The pro-metastatic properties of miR-107 are mediated by repression of its targets, the metastasis suppressors DAPK and KLF4.Citation117 Furthermore, Martello et al showed that miR-107 targets and represses DICER1, a key component of the miRNA processing machinery, thereby attenuating global miRNA production.Citation115 Therefore, elevated levels of miR-107 in tumors may contribute to the global reduction of miRNA abundance that was observed in various cancer types.Citation121 In addition to regulation of mRNA targets, miR-107 can also directly interact with and negatively regulate the let-7 miRNA.Citation116 Accordingly, miR-107 increased the tumorigenic and metastatic potential of human breast cancer cell lines in xenograft mouse models via inhibition of let-7 and upregulation of let-7 targets.Citation116 However, others have shown that miR-107 also has tumor suppressing functions by inhibiting cell proliferation and migration of breast cancer, gastric cancer, and glioma cells.Citation120,Citation122,Citation123 These tumor suppressing effects could be partially attributed to the miR-107-mediated repression of the response to hypoxia and angiogenesis via targeting of HIF 1β, resulting in a decreased supply of oxygen and nutrients and subsequent inhibition of tumor growth.Citation114 The decrease in functional HIF1α–HIF1β dimers after p53-mediated activation of miR-107 may suppress glycolysis under hypoxic conditions. These results indicate that p53-deficient tumors may be resistant to hypoxia not only because of decreased apoptosis and senescence, but also because of increased HIF1 signalling due to the decrease in miR-107 which results in metabolic and angiogenic adaptation. Altogether, the majority of the current data suggests that miR-107 is an oncogenic miRNA that promotes EMT, migration, and metastasis. However, these observations are at first sight not compatible with the induction of miR-107 by p53, which would be expected to mediate tumor suppressive functions. A possible explanation may be that p53 induces miR-107 and thereby downregulates DICER1 to limit the production of p53-induced miRNAs, which would otherwise lead to an unrestrained induction of p53 because of the positive feedback loops these often form with p53.
p53-repressed miRNAs
p53 also directly represses certain miRNAs, including miR-224,Citation124 miR-502,Citation125 and the miR-17-92 cluster.Citation126 However, this type of regulation seems to occur less frequently than the induction of tumor suppressive miRNAs by p53. As expected, miRNAs repressed by p53 mostly have oncogenic functions. The miR-17-92 primary transcript encodes the miRNAs miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1. The miR-17-92 clusters undergo genomic amplification and display elevated expression in various cancer entities, including colon cancer.Citation127 The miRNAs of the miR-17-92 family promote cell proliferation, increase angiogenesis, promote cell survival, and exhibit strong protumorigenic activities in multiple mouse tumor models.Citation128 Yan et al showed that miR-17-92 is repressed upon hypoxia via direct interaction of p53 with the miR-17-92 promoter.Citation126 Due to the strong cell survival promoting properties of miR-17-92 family members, it is likely that the repression of miR-17-92 expression by p53 plays a role in p53-induced apoptosis.
Regulation of p53 by miRNAs
p53 not only regulates the expression and processing of miRNAs, but is also under the control of certain miRNAs. Several miRNAs repress the translation of TP53 mRNA by directly binding to its 3′-UTR ( and ). Since these miRNAs diminish the tumor suppressive activity of p53, they often represent oncomirs. Accordingly, they often exhibit elevated expression in tumors. Similar to the p53-regulated miRNAs the expression of p53-regulating miRNAs is frequently altered in GI tumors ( and ). The first miRNA that was characterized as a direct suppressor of p53 was miR-125b. Le et al. Showed that miR-125b is a negative regulator of p53 expression and p53-induced apoptosis during development and stress response.Citation129 Furthermore, it has been shown that miR-125 targets several additional components of the p53 network, which include both regulators of apoptosis like Bak1, Igfbp3, Itch, Puma, Prkra, Tp53inp1, and Zac1, and components of the cell cycle machinery such as cyclin C, Cdc25c, Cdkn2c, Edn1, Ppp1ca, and Sel1l.Citation130 The authors proposed that by regulating proliferative and apoptotic genes, miR-125b buffers and fine-tunes the activity of the p53 network in order to control the balance between proliferation and apoptosis. Recently, it was shown that the levels of miR-125b are significantly higher in the serum of patients with hepatitis-B-virus-positive HCC and therefore circulating miR-125b may represent a potential non-invasive marker for HCC.Citation131 Moreover, elevated expression of miR-125 was also associated with increased tumor size, enhanced invasion, and poor prognosis in CRC patients.Citation132 Another miRNA that negatively regulates p53 expression via two seed-matching sequences in the human TP53 3′-UTR is miR-504.Citation133 Accordingly, ectopic expression of miR-504 reduced p53 protein levels and impaired p53 functions, especially p53-mediated apoptosis and G1-arrest in response to stress. Furthermore, ectopic expression of miR-504 promoted tumorigenicity of colon cancer cells in mice.Citation133 Additionally, miR-25, miR-30d, miR-33, miR-98, miR-150, miR-214, miR-375, miR-380, and miR-1285 also downregulate p53 protein levels through seed-matching sequences in the 3′-UTR of TP53.Citation134–Citation144 Accordingly, ectopic expression of these miRNAs suppresses p53 expression and induces phenotypes that are consistent with a decrease in p53 function, such as reduced apoptosis and senescence, and increased invasion and stem cell self-renewal.Citation134–Citation137 miR-25 levels were increased in esophageal tumors and serum of esophageal cancer patients displayed elevated levels of circulating miR-25.Citation145 Moreover, expression of miR-25 was significantly higher in colorectal tumors and elevated levels of miR-25 were associated with increased tumor invasion, lymph node metastasis, distant metastasis, TNM (tumor, node, metastasis status based classification) stage, and poor survival of CRC patients.Citation146 MiR-30d is an important regulator of autophagy,Citation147 and interestingly, amplification of the MIR30D gene was found in ∼30% of 1,283 analyzed solid tumors, including bladder, colorectal, and pancreatic cancer.Citation148
In addition to the direct repression of p53 by miRNAs, several miRNAs also regulate the expression of p53 indirectly. As described above, the expression of the p53 inhibitors MDM2 and MDM4 is directly repressed by several p53-induced miRNAs: MDM2 is a target of miR-145, miR-192/194/215, miR-605, and miR-29b, whereas MDM4 is targeted by the miR-34 family members (). Therefore, p53-mediated induction of these miRNAs results in a positive feedback and enhanced p53 activation. In addition, miRNAs of the miR-29 family target the expression of other negative regulators of p53, such as Cdc42, PPM1D, and the regulatory subunit of phosphatidylinositol-3 kinase (PI3K), p85α, and thereby indirectly enhance the levels and activity of p53.Citation149,Citation150 In addition, miR-29 is also directly induced by p53,Citation149 thereby forming a positive feedback loop that is activated during aging and DNA damage, and reinforces p53 effector functions, such as apoptosis and senescence. The members of the miR-29 family are aberrantly expressed in various tumors, including gastric cancer and HCC.Citation151–Citation153 Moreover, low expression of miR-29 in HCC was associated with decreased survival.Citation154 Furthermore, miR-122, which is expressed exclusively in the liver, is frequently downregulated in liver cancer.Citation155 It was shown that miR-122 stabilizes and therefore increases p53 protein levels and activity via downregulation of its target cyclin G1.Citation156 Repression of cyclin G1 results in decreased recruitment of the PP2A phosphatase to the p53-inhibitor MDM2. The resulting decrease in MDM2 activity leads to activation of p53.Citation157 Interestingly, Cyclin G1 is also directly induced by p53 and therefore forms a negative feedback loop with p53.Citation158 However, therapeutic treatment with miR-122 mimtics might abrogate this loop by downregulation of cyclin G1. Indeed, it has been shown that ectopic miR-122 expression increases the sensitivity of HCC cell lines to the chemotherapeutic agent doxorubicin, which is known to induce p53.Citation156 However, it should be noted that miR-122 increases chemosensitivity also in the absence of wild-type p53 and therefore seems to have p53-independent functions.
Conclusion and outlook
Although numerous links between p53 and miRNAs have been identified, we have only begun to understand the complex interplay between p53 and miRNAs in tumor suppression. Therefore, additional efforts are necessary to uncover more details of the p53/miRNA network. Since p53 has many functions in tumor suppression, future research should focus on identifying which miRNA is responsible for mediating specific p53 functions. It would also be of interest to investigate whether the complete spectrum of tumor suppressing functions of p53, which is frequently lost in tumors, can be restored by the introduction of specific combinations of p53-induced miRNAs. So far, the majority of studies have rather focused on the identification and characterization of single p53-regulated miRNAs. A feasible strategy for a comprehensive, genome-wide identification of p53-regulated miRNAs and their target genes has been recently described by us.Citation159 This strategy employs a combination of various unbiased genome-wide next generation sequencing screens to simultaneously identify and characterize p53-regulated miRNAs and their targets. By now, knockout mouse models of single p53-induced miRNAs have not fully recapitulated the cancerous phenotype of p53 knockout mice, which is characterized by the early onset of lymphomas.Citation160 Therefore, it would be interesting to generate genetically modified mice that simultaneously lack multiple p53-related miRNAs to investigate whether loss of certain combinations of miRNAs fully or at least partially mimics the phenotype of p53-knockout mice. Complementarily, p53 knockout mice could be treated with a cocktail of p53-induced miRNAs to investigate whether certain combinations of miRNAs can suppress the tumor promoting effects of p53 loss. In light of the central role of p53-regulated miRNAs or p53-regulating miRNAs for tumor suppression, the introduction of these miRNAs or of miRNA antagonists into tumor cells represents an exciting possibility for novel cancer-therapeutic approaches. Such therapies could also be performed in combination with standard anticancer therapies as has been already shown for miR-34a, which sensitized gastric cancer cells to the chemotherapeutic drugs docetaxel, gemcitabine, cisplatin, and doxorubicin.Citation63 A major obstacle for such approaches is the currently low efficiency of miRNA delivery into tumor cells. Therefore, further research is needed to develop more efficient strategies for in vivo miRNA delivery. Promisingly, improved delivery using novel nanoparticles was recently employed to show that a combination of miR-34a mimics and siRNAs directed at mutant oncogenes is more effective than either RNA alone in a pre-clinical mouse model of lung cancer.Citation161 Since p53-regulated miRNAs are often inactivated by CpG methylation in tumors, establishment of routine protocols for detection of methylation of selected promoters of miRNA-encoding genes in body fluids may represent an important aspect of future GI cancer diagnostics.
Disclosure
The authors report no conflicts of interest in this work.
References
- International Agency for Research on CancerGLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012GenevaWorld Health Organization2014 Available from: http://globocan.iarc.fr/Pages/fact_sheets_population.aspxAccessed July 30, 2014
- International Agency for Research on CancerIARC TP53 DatabseGenevaWorld Health Organization2014 Available from: http://p53.iarc.fr/TP53SomaticMutations.aspxAccessed July 30, 2014
- VogelsteinBLaneDLevineAJSurfing the p53 networkNature2000408681030731011099028
- OrenMDecision making by p53: life, death and cancerCell Death Differ200310443144212719720
- GohAMCoffillCRLaneDPThe role of mutant p53 in human cancerJ Pathol2011223211612621125670
- LaneDLevineAp53 Research: the past thirty years and the next thirty yearsCold Spring Harb Perspect Biol2010212a00089320463001
- el-DeiryWSKernSEPietenpolJAKinzlerKWVogelsteinBDefinition of a consensus binding site for p53Nat Genet19921145491301998
- HermekingHp53 enters the microRNA worldCancer Cell200712541441817996645
- HermekingHMicroRNAs in the p53 network: micromanagement of tumour suppressionNat Rev Cancer201212961362622898542
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- KrolJLoedigeIFilipowiczWThe widespread regulation of microRNA biogenesis, function and decayNat Rev Genet201011959761020661255
- YatesLANorburyCJGilbertRJThe long and short of microRNACell2013153351651923622238
- FriedmanRCFarhKKBurgeCBBartelDPMost mammalian mRNAs are conserved targets of microRNAsGenome Res20091919210518955434
- FassanMCroceCMRuggeMmiRNAs in precancerous lesions of the gastrointestinal tractWorld J Gastroenterol201117485231523922219591
- SongBJuJImpact of miRNAs in gastrointestinal cancer diagnosis and prognosisExpert Rev Mol Med201012e3320942990
- BoniVBandresEZarateRColucciGMaielloEGarcia-FoncillasJMicroRNAs as a new potential therapeutic opportunity in gastrointestinal cancerOncology200977Suppl 1758920130435
- VicentiniCFassanMD’AngeloEClinical application of microRNA testing in neuroendocrine tumors of the gastrointestinal tractMolecules20141922458246824566314
- MachaMASeshacharyuluPKrishnSRMicroRNAs (miRNA) as Biomarker(s) for Prognosis and Diagnosis of Gastrointestinal (GI) CancersCurr Pharm Des Epub January 28, 2014
- AjitSKCirculating microRNAs as biomarkers, therapeutic targets, and signaling moleculesSensors (Basel)20121233359336922737013
- Blanco-CalvoMCalvoLFigueroaAHaz-CondeMAntón-AparicioLValladares-AyerbesMCirculating microRNAs: molecular microsensors in gastrointestinal cancerSensors (Basel)20121279349936223012546
- BommerGTGerinIFengYp53-mediated activation of miRNA34 candidate tumor-suppressor genesCurr Biol200717151298130717656095
- ChangTCWentzelEAKentOATransactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosisMol Cell200726574575217540599
- CorneyDCFlesken-NikitinAGodwinAKWangWNikitinAYMicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growthCancer Res200767188433843817823410
- HeLHeXLimLPA microRNA component of the p53 tumour suppressor networkNature200744771481130113417554337
- Raver-ShapiraNMarcianoEMeiriETranscriptional activation of miR-34a contributes to p53-mediated apoptosisMol Cell200726573174317540598
- TarasovVJungPVerdoodtBDifferential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrestCell Cycle20076131586159317554199
- TazawaHTsuchiyaNIzumiyaMNakagamaHTumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cellsProc Natl Acad Sci U S A200710439154721547717875987
- SuzukiHIYamagataKSugimotoKIwamotoTKatoSMiyazonoKModulation of microRNA processing by p53Nature2009460725452953319626115
- MudhasaniRZhuZHutvagnerGLoss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cellsJ Cell Biol200818171055106318591425
- SuXChakravartiDChoMSTAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAsNature2010467731898699020962848
- LéveilléNElkonRDavalosVSelective inhibition of microRNA accessibility by RBM38 is required for p53 activityNat Commun2011251322027593
- KimTVeroneseAPichiorriFp53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2J Exp Med2011208587588321518799
- FabbriMBottoniAShimizuMAssociation of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemiaJAMA20113051596721205967
- BraunCJZhangXSavelyevaIp53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrestCancer Res20086824100941010419074875
- SachdevaMZhuSWuFp53 represses c-Myc through induction of the tumor suppressor miR-145Proc Natl Acad Sci U S A200910693207321219202062
- BöhligLFriedrichMEngelandKp53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteinsNucleic Acids Res201139244045320833636
- WangJHeQHanCp53-facilitated miR-199a-3p regulates somatic cell reprogrammingStem Cells20123071405141322553189
- RoySLeviEMajumdarAPSarkarFHExpression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDFJ Hematol Oncol201255822992310
- CuiXZhaoZLiuDInactivation of miR-34a by aberrant CpG methylation in Kazakh patients with esophageal carcinomaJ Exp Clin Cancer Res2014332024528540
- StánitzEJuhászKTóthCGombosKNataliPGEmberIEvaluation of MicroRNA expression pattern of gastric adenocarcinoma associated with socioeconomic, environmental and lifestyle factors in northwestern HungaryAnticancer Res20133383195320023898078
- XieKLiuJChenJMethylation-associated silencing of microRNA-34b in hepatocellular carcinoma cancerGene2014543110110724704024
- WangRMaJWuQFunctional role of miR-34 family in human cancerCurr Drug Targets201314101185119123834144
- RokavecMLiHJiangLHermekingHThe p53/miR-34 axis in development and diseaseJ Mol Cell Biol20146321423024815299
- MandkePWyattNFraserJBatesBBerberichSJMarkeyMPMicroRNA-34a modulates MDM4 expression via a target site in the open reading framePLoS One201278e4203422870278
- OkadaNLinCPRibeiroMCA positive feedback between p53 and miR-34 miRNAs mediates tumor suppressionGenes Dev201428543845024532687
- YamakuchiMFerlitoMLowensteinCJmiR-34a repression of SIRT1 regulates apoptosisProc Natl Acad Sci U S A200810536134211342618755897
- ChoiSEFuTSeokSElevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPTAging Cell20131261062107223834033
- MenssenAHydbringPKapelleKThe c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loopProc Natl Acad Sci U S A20121094E187E19622190494
- MarshallGMLiuPYGherardiSSIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stabilityPLoS Genet201176e100213521698133
- SiemensHJackstadtRHüntenSmiR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitionsCell Cycle201110244256427122134354
- BurkUSchubertJWellnerUA reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cellsEMBO Rep20089658258918483486
- GregoryPABertAGPatersonELThe miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1Nat Cell Biol200810559360118376396
- AhnYHGibbonsDLChakravartiDZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expressionJ Clin Invest201212293170318322850877
- HahnSJackstadtRSiemensHHüntenSHermekingHSNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transitionEMBO J201332233079309524185900
- RokavecMÖnerMGLiHIL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasisJ Clin Invest201412441853186724642471
- BaderAGmiR-34 – a microRNA replacement therapy is headed to the clinicFront Genet2012312022783274
- PramanikDCampbellNRKarikariCRestitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in miceMol Cancer Ther20111081470148021622730
- CraigVJTzankovAFloriMSchmidCABaderAGMüllerASystemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivoLeukemia201226112421242422522790
- WigginsJFRuffinoLKelnarKDevelopment of a lung cancer therapeutic based on the tumor suppressor microRNA-34Cancer Res201070145923593020570894
- Mirna Therapeutics, Inc.homepage on the InternetAustinMirna Therapeutics, Inc.2014 Available from: http://www.mirnarx.com/Accessed April 30, 2014
- FujitaYKojimaKHamadaNEffects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cellsBiochem Biophys Res Commun2008377111411918834855
- VinallRLRipollAZWangSPanC-XdeVere WhiteRWMiR-34a chemosensitizes bladder cancer cells to cisplatin treatment regardless of p53-Rb pathway statusInt J Cancer2012130112526253821702042
- JiQHaoXMengYRestoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheresBMC Cancer2008826618803879
- JiQHaoXZhangMMicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cellsPLoS One200948e681619714243
- SiemensHJackstadtRKallerMHermekingHRepression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemnessOncotarget2013491399141524009080
- LodyginDTarasovVEpanchintsevAInactivation of miR-34a by aberrant CpG methylation in multiple types of cancerCell Cycle20087162591260018719384
- VogtMMundingJGrünerMFrequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomasVirchows Arch2011458331332221225432
- ToyotaMSuzukiHSasakiYEpigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancerCancer Res200868114123413218519671
- SuzukiHYamamotoENojimaMMethylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defectCarcinogenesis201031122066207320924086
- SiemensHNeumannJJackstadtRDetection of miR-34a promoter methylation in combination with elevated expression of c-Met and β-catenin predicts distant metastasis of colon cancerClin Cancer Res201319371072023243217
- HermekingHThe miR-34 family in cancer and apoptosisCell Death Differ201017219319919461653
- KongDHeathEChenWEpigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatmentAm J Transl Res201241142322347519
- ÖstlingPLeivonenSKAakulaASystematic analysis of microRNAs targeting the androgen receptor in prostate cancer cellsCancer Res20117151956196721343391
- WuXDSongYCCaoPLDetection of miR-34a and miR-34b/c in stool sample as potential screening biomarkers for noninvasive diagnosis of colorectal cancerMed Oncol201431489424573638
- CalinGADumitruCDShimizuMFrequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemiaProc Natl Acad Sci U S A20029924155241552912434020
- KleinULiaMCrespoMThe DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemiaCancer Cell2010171284020060366
- QianJJiangBLiMChenJFangMPrognostic significance of microRNA-16 expression in human colorectal cancerWorld J Surg201337122944294924045965
- ZhangXJYeHZengCWHeBZhangHChenYQDysregulation of miR-15a and miR-214 in human pancreatic cancerJ Hematol Oncol201034621106054
- MaQWangXLiZmicroRNA-16 represses colorectal cancer cell growth in vitro by regulating the p53/survivin signaling pathwayOncol Rep20132941652165823380758
- DaiLWangWZhangSVector-based miR-15a/16-1 plasmid inhibits colon cancer growth in vivoCell Biol Int201236876577022574716
- WuGYuFXiaoZHepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitroBr J Cancer2011105114615321629246
- CimminoACalinGAFabbriMmiR-15 and miR-16 induce apoptosis by targeting BCL2Proc Natl Acad Sci U S A200510239139441394916166262
- LiuQFuHSunFmiR-16 family induces cell cycle arrest by regulating multiple cell cycle genesNucleic Acids Res200836165391540418701644
- BonciDCoppolaVMusumeciMThe miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activitiesNat Med200814111271127718931683
- ShiLJackstadtRSiemensHLiHKirchnerTHermekingHp53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancerCancer Res201474253254224285725
- JackstadtRRöhSNeumannJAP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancerJ Exp Med201321071331135023752226
- SuzukiKMatsubaraHRecent advances in p53 research and cancer treatmentJ Biomed Biotechnol2011201197831221765642
- KanoMSekiNKikkawaNmiR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinomaInt J Cancer2010127122804281421351259
- GaoPXinga-YZhouGYThe molecular mechanism of microRNA-145 to suppress invasion-metastasis cascade in gastric cancerOncogene201332449150122370644
- PapaconstantinouIGMantaAGazouliMExpression of microR-NAs in patients with pancreatic cancer and its prognostic significancePancreas2013421677122850622
- SlabyOSvobodaMFabianPAltered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancerOncology2007725–639740218196926
- RatertNMeyerHAJungMmiRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcomeJ Mol Diagn201315569570523945108
- PagliucaaValvoCFabriziEAnalysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repressionOncogene201332404806481323128394
- IbrahimAFWeirauchUThomasMGrünwellerAHartmannRKAignerAMicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinomaCancer Res201171155214522421690566
- RenDWangMGuoWWild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR145Int J Oncol20134241473148123404342
- KentOAChivukulaRRMullendoreMRepression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathwayGenes Dev201024242754275921159816
- XuNPapagiannakopoulosTPanGThomsonJaKosikKSMicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cellsCell2009137464765819409607
- KrizhanovskyVLoweSWStem cells: The promises and perils of p53Nature200946072591085108619713919
- ZhangJSunQZhangZGeSHanZGChenWTLoss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loopOncogene2013321616922330136
- PichiorriFSuhS-sRocciADownregulation of p53-inducible microRNAs 192, 194,and 215 Impairs the p53/MDM2 Auto-regulatory Loop in Multiple Myeloma DevelopmentCancer Cell201018436738120951946
- LiSGaoJGuJYuanJHuaDShenLMicroRNA-215 inhibits relapse of colorectal cancer patients following radical surgeryMed Oncol201330254923532818
- GeorgesSABieryMCKimSYCoordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215Cancer Res20086824101051011219074876
- GengLChaudhuriATalmonGMicroRNA-192 suppresses liver metastasis of colon cancerOncogene Epub November 11, 2013
- DongPKaneuchiMWatariHMicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1Mol Cancer2011109921851624
- WellnerUSchubertJBurkUCThe EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAsNat Cell Biol200911121487149519935649
- ShimonoYZabalaMChoRWDownregulation of miRNA-200c links breast cancer stem cells with normal stem cellsCell2009138359260319665978
- LuYXYuanLXueXLRegulation of colorectal carcinoma stemness, growth and metastasis by a miR-200-c-Sox2 negative feedback loop mechanismClin Cancer Res201420102631264224658157
- TanakaKMiyataHYamasakiMCirculating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancerAnn Surg Oncol201320Suppl 3S607S61523838916
- ToiyamaYHurKTanakaKSerum miR-200c Is a Novel Prognostic and Metastasis-Predictive Biomarker in Patients With Colorectal CancerAnn Surg2014259473574323982750
- Valladares-AyerbesMReboredoMMedina-VillaamilVCirculating miR-200c as a diagnostic and prognostic biomarker for gastric cancerJ Transl Med20121018622954417
- YuHDuanBJiangLSerum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapyAm J Transl Res201361717724349623
- ZhangGJZhouTLiuZLTianHPXiaSSPlasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinomaMol Clin Oncol20131237938424649179
- TsaiJHDonaherJLMurphyDAChauSYangJSpatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasisCancer Cell201222672573623201165
- YamakuchiMLottermanCDBaoCP53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesisProc Natl Acad Sci U S A2010107146334633920308559
- MartelloGRosatoAFerrariFA MicroRNA targeting dicer for metastasis controlCell201014171195120720603000
- ChenPSSuJLChaSTmiR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humansJ Clin Invest201112193442345521841313
- ChenHYLinYMChungHCmiR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4Cancer Res201272143631364122593189
- LiXZhangYShiYMicroRNA-107, an oncogene microRNA that regulates tumour invasion and metastasis by targeting DICER1 in gastric cancerJ Cell Mol Med20111591887189521029372
- InoueTIinumaHOgawaEInabaTFukushimaRClinicopathological and prognostic significance of microRNA-107 and its relationship to DICER1 mRNA expression in gastric cancerOncol Rep20122761759176422407237
- FengLXieYZhangHWuYmiR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cellsMed Oncol201229285686321264532
- KumarMSLuJMercerKLGolubTRJacksTImpaired microRNA processing enhances cellular transformation and tumorigenesisNat Genet200739567367717401365
- LiFLiuBGaoYUpregulation of MicroRNA-107 induces proliferation in human gastric cancer cells by targeting the transcription factor FOXO1FEBS Lett2014588453854424374340
- ChenLChenXRChenFFMicroRNA-107 inhibits U87 glioma stem cells growth and invasionCell Mol Neurobiol201333565165723572380
- LiangMYaoGYinMTranscriptional cooperation between p53 and NF-κB p65 regulates microRNA-224 transcription in mouse ovarian granulosa cellsMol Cell Endocrinol20133701–211912923474441
- ZhaiHSongBXuXZhuWJuJInhibition of autophagy and tumor growth in colon cancer by miR-502Oncogene201332121570157922580605
- YanHLXueGMeiQRepression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosisEMBO J200928182719273219696742
- TsuchidaAOhnoSWuWmiR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancerCancer Sci2011102122264227121883694
- OliveVJiangIHeLmir-17-92, a cluster of miRNAs in the midst of the cancer networkInt J Biochem Cell Biol20104281348135420227518
- LeMTTehCShyh-ChangNMicroRNA-125b is a novel negative regulator of p53Genes Dev200923786287619293287
- LeMTShyh-ChangNKhawSLConserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairsPLoS Genet201179e100224221935352
- GirayBGEmekdasGTezcanSProfiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinomaMol Biol Rep20144174513451924595450
- NishidaNYokoboriTMimoriKMicroRNA miR-125b is a prognostic marker in human colorectal cancerInt J Oncol20113851437144321399871
- HuWChanCSWuRNegative regulation of tumor suppressor p53 by microRNA miR-504Mol Cell201038568969920542001
- KumarMLuZTakwiAANegative regulation of the tumor suppressor p53 gene by microRNAsOncogene201130784385320935678
- Herrera-MerchanACerratoCLuengoGmiR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewalCell Cycle20109163277328520703086
- SwarbrickAWoodsSLShawAmiR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastomaNat Med201016101134114020871609
- XuXChenZZhaoXMicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinomaBiochem Biophys Res Commun2012421464064522450326
- ZhangNWeiXXuLmiR-150 promotes the proliferation of lung cancer cells by targeting P53FEBS Lett2013587152346235123747308
- LiuYXingRZhangXmiR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cellsDNA Repair (Amst)201312974175023835407
- TianSHuangSWuSGuoWLiJHeXMicroRNA-1285 inhibits the expression of p53 by directly targeting its 3′ untranslated regionBiochem Biophys Res Commun2010396243543920417621
- XuCXXuMTanLMicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/NanogJ Biol Chem201228742349703497822927443
- ZhangYGaoJSTangXMicroRNA 125a and its regulation of the p53 tumor suppressor geneFEBS Lett2009583223725373019818772
- ZhangSZhangCLiYWangPYueZXieSmiR-98 regulates cisplatin-induced A549 cell death by inhibiting TP53 pathwayBiomed Pharmacother201165643644221880462
- WangDTMaZLLiYLmiR-150, p53 protein and relevant miRNAs consist of a regulatory network in NSCLC tumorigenesisOncol Rep201330149249823670238
- WuCLiMHuCDuanHClinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinomaMol Biol Rep20144131257126624390317
- LiXYangCWangXZhangJZhangRLiuRThe expression of miR-25 is increased in colorectal cancer and is associated with patient prognosisMed Oncol201431178124293092
- YangXZhongXTanyiJLmir-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cellsBiochem Biophys Res Commun2013431361762223274497
- LiNKaurSGreshockJA combined array-based comparative genomic hybridization and functional library screening approach identifies mir-30d as an oncomir in cancerCancer Res201272115416422058146
- UgaldeAPRamsayAJde la RosaJAging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53EMBO J201130112219223221522133
- ParkSYLeeJHHaMNamJWKimVNmiR-29 miRNAs activate p53 by targeting p85 alpha and CDC42Nat Struct Mol Biol2009161232919079265
- WangYZhangXLiHYuJRenXThe role of miRNA-29 family in cancerEur J Cell Biol201392312312823357522
- GongJLiJWangYCharacterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancerCarcinogenesis201435249750624130168
- ZhuH-TDongQ-ZShengY-YMicroRNA-29a-5p is a novel predictor for early recurrence of hepatitis B virus-related hepatocellular carcinoma after surgical resectionPLoS One2012712e5239323285022
- XiongYFangJHYunJPEffects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinomaHepatology201051383684520041405
- GramantieriLFornariFCallegariEMicroRNA involvement in hepatocellular carcinomaJ Cell Mol Med2008126A2189220419120703
- FornariFGramantieriLGiovanniniCMiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cellsCancer Res200969145761576719584283
- OkamotoKLiHJensenMRCyclin G recruits PP2A to dephosphorylate Mdm2Mol Cell20029476177111983168
- OkamotoKBeachDCyclin G is a transcriptional target of the p53 tumor suppressor proteinEMBO J19941320481648227957050
- HüntenSSiemensHKallerMHermekingHThe p53/microRNA network in cancer: experimental and bioinformatics approachesAdv Exp Med Biol20137747710123377969
- ConcepcionCPHanYCMuPIntact p53-dependent responses in miR-34-deficient micePLoS Genet201287e100279722844244
- XueWDahlmanJETammelaTSmall RNA combination therapy for lung cancerProc Natl Acad Sci USA201411134E3553356125114235
- NugentMMillerNKerinMJCirculating miR-34a levels are reduced in colorectal cancerJ Surg Oncol2012106894795222648208
- WuJWuGLvLMicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1Carcinogenesis201233351952822198213
- ChenXHuHGuanXCpG island methylation status of miRNAs in esophageal squamous cell carcinomaInt J Cancer201213071607161321547903
- JamiesonNBMorranDCMortonJPMicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinomaClin Cancer Res201218253454522114136
- HarataKIshiguroHKuwabaraYKimuraMMitsuiAMicroRNA-34b has an oncogenic role in esophageal squamous cell carcinomaOncol Lett20101468568922966364
- LiuSGQinXGZhaoBSDifferential expression of miRNAs in esophageal cancer tissueOncol Lett2013551639164223761828
- DiazTTejeroRMorenoIRole of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidinesJ Surg Oncol2014109767668324510588
- HurKToiyamaYTakahashiMMicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasisGut20136291315132622735571
- ChenJWangWZhangYChenYHuTPredicting distant metastasis and chemoresistance using plasma miRNAsMed Oncol201431179924310813
- DavalosVMoutinhoCVillanuevaADynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesisOncogene201231162062207421874049
- DuYXuYDingLDown-regulation of miR-141 in gastric cancer and its involvement in cell growthJ Gastroenterol200944655656119363643
- KarakatsanisAPapaconstantinouIGazouliMLyberopoulouAPolymeneasGVorosDExpression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significanceMol Carcinog201352429730322213236
- YehTSWangFChenTCExpression profile of microRNA-200 family in hepatocellular carcinoma with bile duct tumor thrombusAnn Surg2014259234635424135722
- SunYShenSLiuXMiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinomaMol Cell Biochem20143901-2193024402783
- ChenZSaadRJiaPGastric adenocarcinoma has a unique microRNA signature not present in esophageal adenocarcinomaCancer2013119111985199323456798
- LiAOmuraNHongSMPancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levelsCancer Res201070135226523720551052
- SharmaPSarayaAGuptaPSharmaRDecreased levels of circulating and tissue miR-107 in human esophageal cancerBiomarkers201318432233023627613
- LeeKHLottermanCKarikariCEpigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancerPancreatology20099329330119407485
- FengYZhuJOuCMicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1Br J Cancer201411092300230924642628
- EarleJSLuthraRRomansAAssociation of microRNA expression with microsatellite instability status in colorectal adenocarcinomaJ Mol Diagn201012443344020413677
- LiJMZhaoRHLiSTDown-regulation of fecal miR-143 and miR-145 as potential markers for colorectal cancerSaudi Med J2012331242922273643
- QiuTZhouXWangJMiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancerFEBS Lett201458871168117724613927
- XingAYWangBShiDBDeregulated expression of miR-145 in manifold human cancer cellsExp Mol Pathol2013951919723714355
- ChiangYSongYWangZmicroRNA-192, -194 and -215 are frequently downregulated in colorectal cancerExp Ther Med20123356056622969930
- FaltejskovaPSvobodaMSrutovaKIdentification and functional screening of microRNAs highly deregulated in colorectal cancerJ Cell Mol Med201216112655266622469014
- KahlertCKluppFBrandKInvasion front-specific expression and prognostic significance of microRNA in colorectal liver metastasesCancer Sci2011102101799180721722265
- SongYZhaoFWangZInverse association between miR-194 expression and tumor invasion in gastric cancerAnn Surg Oncol201219Suppl 3S509S51721845495
- DengYHuangZXuYMiR-215 modulates gastric cancer cell proliferation by targeting RB1Cancer Lett20143421273523981575
- WangLGGuJSerum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasisCancer Epidemiol2012361e61e6722018950
- TangWZhuYGaoJMicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4Br J Cancer2014110245045824281002
- ChenLXiaoHWangZ-HmiR-29a suppresses growth and invasion of gastric cancer cells in vitro by targeting VEGF-ABMB Rep2014471394424209632
- XiaoJLinHLuoXLuoXWangZmiR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stressEMBO J201130352453221217645
- JinLHuWLJiangCCMicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanomaProc Natl Acad Sci U S A201110838158401584521896753
- ØsterBLinnetLChristensenLLCOLOFOL steering groupNon-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancerInt J Cancer2013132102303231523115050
- WangFMaYLZhangPSP1 mediates the link between methy-lation of the tumour suppressor miR-149 and outcome in colorectal cancerJ Pathol20132291122422821729
- WangYZhengXZhangZMicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancerPLoS One2012710e4169323144691
- LinJHuoRXiaoLA novel p53/microRNA-22/Cyr61 axis in synovial cells regulates inflammation in rheumatoid arthritisArthritis Rheumatol2014661495924449575
- ZhangGXiaSTianHLiuZZhouTClinical significance of miR-22 expression in patients with colorectal cancerMed Oncol20122953108311222492279
- ZhangCWangCChenXExpression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinomaClin Chem201056121871187920943850
- GanepolaGARutledgeJRSumanPYiengpruksawanAChangDHNovel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancerWorld J Gastrointest Oncol201461223324578785
- GuoMMHuLHWangYQmiR-22 is down-regulated in gastric cancer, and its overexpression inhibits cell migration and invasion via targeting transcription factor Sp1Med Oncol201330254223529765
- WangWLiFZhangYTuYYangQGaoXReduced expression of miR-22 in gastric cancer is related to clinicopathologic characteristics or patient prognosisDiagn Pathol2013810223786758
- ZhouLHeJZhangYMicroRNA-22 expression in hepatocellular carcinoma and its correlation with ezrin proteinJ Int Med Res20134141009101623766411
- ZhangJYangYYangTmicroRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicityBr J Cancer201010381215122020842113
- Au YeungCLTsangTYYauPLKwokTTHuman papillomavirus type 16 E6 induces cervical cancer cell migration through the p53/microRNA-23b/urokinase-type plasminogen activator pathwayOncogene201130212401241021242962
- ZhaoBSLiuSGWangTYScreening of microRNA in patients with esophageal cancer at same tumor node metastasis stage with different prognosesAsian Pac J Cancer Prev201314113914323534712
- SalviASabelliCMonciniSMicroRNA-23b mediates uroki-nase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cellsFEBS J2009276112966298219490101
- ZhangHHaoYYangJGenome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasisNat Commun2011255422109528
- PiovanCPalmieriDDi LevaGOncosuppressive role of p53-induced miR-205 in triple negative breast cancerMol Oncol20126445847222578566
- GuJWangYWuXMicroRNA in the pathogenesis and prognosis of esophageal cancerCurr Pharm Des20131971292130023092349
- SaadRChenZZhuSDeciphering the unique microRNA signature in human esophageal adenocarcinomaPLoS One201385e6446323724052
- ZhangTZhangJCuiMHepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesisNeoplasia201315111282129124339740
- ZhangYLiaoJMZengSXLuHp53 downregulates Down syndrome-associated DYRK1A through miR-1246EMBO Rep201112881181721637297
- PiepoliATavanoFCopettiMMirna expression profiles identify drivers in colorectal and pancreatic cancersPLoS One201273e3366322479426
- Ogata-KawataHIzumiyaMKuriokaDCirculating exosomal microRNAs as biomarkers of colon cancerPLoS One201494e9292124705249
- FuHLWu dePWangXFAltered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, ChinaCell Biochem Biophys201367265766823516093
- TakeshitaNHoshinoIMoriMSerum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinomaBr J Cancer2013108364465223361059
- BarsottiAMBeckermanRLaptenkoOHuppiKCaplenNJPrivesCp53-Dependent induction of PVT1 and miR-1204J Biol Chem201228742509251922110125
- KogaYYasunagaMTakahashiAMicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screeningCancer Prev Res (Phila)20103111435144220959518
- DiosdadoBvan de WielMATerhaar Sive DrosteJSMiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progressionBr J Cancer2009101470771419672269
- XuXLJiangYHFengJGSuDChenPCMaoWMMicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinomaAnn Thorac Surg20149731037104524360091
- LiuMWangZYangSTNF-alpha is a novel target of miR-19aInt J Oncol20113841013102221271217
- TsujiuraMKomatsuSIchikawaDCirculating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancerGastric Cancer Epub March 14, 2014
- ShiotaniAUedoNIishiHH. pylori eradication did not improve dysregulation of specific oncogenic miRNAs in intestinal metaplastic glandsJ Gastroenterol201247998899822382634
- ShigokaMTsuchidaAMatsudoTDeregulation of miR-92a expression is implicated in hepatocellular carcinoma developmentPathol Int201060535135720518884
- MorimuraRKomatsuSIchikawaDNovel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancerBr J Cancer2011105111733174022045190
- ZhangGJZhouHXiaoHXLiYZhouTUp-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancerCancer Cell Int201313110424152489
- LiaoWTLiTTWangZGmicroRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2Clin Cancer Res201319174662467223846336
- YuanKXieKFoxJDecreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in miceGastroenterology20131454853864.e923770133
- SciscianiCVossioSGuerrieriFTranscriptional regulation of miR-224 upregulated in human HCCs by NFkappaB inflammatory pathwaysJ Hepatol201256485586122178270
- ZhangYTakahashiSTasakaAYoshimaTOchiHChayamaKInvolvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinomaJ Gastroenterol Hepatol201328356557522989374
- KomatsuSIchikawaDHirajimaSPlasma microRNA profiles: identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinomaBr J Cancer2014
- ZhuCRenCHanJA five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancerBr J Cancer201411092291229924595006
- ZhaoHWangYYangLJiangRLiWMiR-25 promotes gastric cancer cells growth and motility by targeting RECKMol Cell Biochem20143851–220721324078004
- UedaTVoliniaSOkumuraHRelation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysisLancet Oncol201011213614620022810
- KimBHHongSWKimAChoiSHYoonSOPrognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinomaJ Surg Oncol2013107550551022996433
- YaoJLiangLHuangSMicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinomaHepatology201051384685620054866
- HuangS-DYuanYZhuangC-WMicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinomaMol Cancer2012115122867052
- FassanMPizziMRealdonSThe HER2-miR125a5p/miR125b loop in gastric and esophageal carcinogenesisHum Pathol20134491804181023618359
- NishidaNMimoriKFabbriMMicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumabClin Cancer Res20111792725273321220473
- HashiguchiYNishidaNMimoriKDown-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significanceInt J Oncol20124051477148222322911
- BiQTangSXiaLEctopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGFPLoS One201276e4016922768249
- KimJKNohJHJungKHSirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125bHepatology20135731055106723079745
- LiangLWongC-MYingQMicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2Hepatology20105251731174020827722
- BloomstonMFrankelWLPetroccaFMicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitisJAMA2007297171901190817473300
- MaYZhangPWangFmiR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancerGut201261101447145322052060
- YokoboriTSuzukiSTanakaNMiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1Cancer Sci20131041485423013135
- WuQJinHYangZMiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2Biochem Biophys Res Commun2010392334034520067763
- SrivastavaSKBhardwajASinghSMicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cellsCarcinogenesis201132121832183921983127
- ZhouYHongLPrediction value of miR-483 and miR-214 in prognosis and multidrug resistance of esophageal squamous cell carcinomaGenet Test Mol Biomarkers201317647047423721345
- YangTSYangXHWangXDWangYLZhouBSongZSMiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTENCancer Cell Int20131316823834902
- ShihTCTienYJWenCJMicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatomaJ Hepatol201257358459122613005
- XiaHOoiLLHuiKMMiR-214 targets β-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinomaPLoS One201279e4420622962603
- WangJLiJWangXZhengCMaWDownregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasisBiochem Biophys Res Commun20134391475323962428
