Abstract
Long noncoding RNAs (lncRNAs) are typically defined as transcripts longer than 200 nucleotides. lncRNAs can regulate gene expression at epigenetic, transcriptional, and posttranscriptional levels. Recent studies have shown that lncRNAs are involved in many neurological diseases such as epilepsy, neurodegenerative conditions, and genetic disorders. Alzheimer’s disease is a neurodegenerative disease, which accounts for >80% of dementia in elderly subjects. In this review, we will highlight recent studies investigating the role of lncRNAs in Alzheimer’s disease and focus on some specific lncRNAs that may underlie Alzheimer’s disease pathophysiology and therefore could be potential therapeutic targets.
Introduction
Alzheimer’s disease (AD) is one of the most common neurodegenerative disease and accounts for >80% of dementia cases in people aged older than 65 years.Citation1 The disease is characterized by devastating symptoms such as apraxia, agnosia, aphasia, and emotional disturbance because of progressive mental and behavioral function decline.Citation2–Citation4 The 2015 Alzheimer’s Association report predicts that, by 2050, there will be a new diagnostic case every 33 seconds, corresponding to 1 million new AD patients every year.Citation5 Given the disability and dependence of these patients, the increasing prevalence of AD will impose huge burdens on families and society. Long noncoding RNAs (lncRNAs) comprise a subgroup of noncoding RNAs (ncRNAs) longer than 200 nucleotides (nt), accounting for the largest proportion of the mammalian noncoding transcriptome. lncRNAs impact AD pathogenesis because of their diverse biochemical and functional effects such as chromatin modulation, posttranscriptional and post-translational regulation, and protein complex organization.Citation6,Citation7
AD pathophysiology
Since the time of Dr Alois Alzheimer, neuropathologists have known that brain tissue of patients with AD contains extracellular senile plaques and intracellular neurofibrillary tangles composed of amyloid beta (Aβ) protein and hyperphosphorylated tau protein, respectively.Citation8–Citation15 Amyloid precursor protein (APP) is sequentially cleaved by β-site APP cleaving enzyme-1 (BACE1), and γ-secretase during Aβ biosynthesis, with γ-secretase initiating the “amyloid-cascade”.Citation16 Aβ peptides aggregate into soluble oligomers that are proposed to be the activator of N-methyl-d aspartate receptor endocytosis, mitochondrial dysfunction, oxidative damage, excessive calcium influx, lipid dysregulation, synaptic dysfunction, neuronal stress, apoptosis, aberrant neurogenesis, and neuroinflammation. However, whether or not Aβ induces tau aggregation is still being debated.Citation17–Citation21 But most recent studies suggest that Aβ oligomer formation may be the essential step in the pathophysiology underpinnings of AD.Citation17,Citation22–Citation24
lncRNA
ncRNAs can be divided on the basis of size into short ncRNAs (<200 nt in length) and lncRNAs.Citation17,Citation25 lncRNAs vary from 200 nt to over 100 kb and usually lack an obvious open reading frame.Citation26–Citation30 lncRNAs secondary structure connected to specific functions are evolutionarily conserved.Citation31,Citation32 They regulate dynamically, localizing at specific cell types and in subcellular compartments.Citation26,Citation33,Citation34 lncRNAs regulate gene expression at different levels.Citation35 Most lncRNAs are located in the nucleus, which is consistent with their major function of epigenetic regulation.Citation26,Citation36 lncRNAs are not considered to be the “dark matter”, rather they have essential roles in controlling transcription and translation, as well as during genetic imprinting, genome rearrangement, chromatin modification regulation of the cell cycle, transcription, splicing, messenger RNA (mRNA) decay, and translation.Citation27,Citation30,Citation37 The pathomechanism and genetic factors of AD have been investigated for nearly 100 years. Research is ongoing, many studies have demonstrated that dysregulation of lncRNAs involved in cancer; epilepsy; and cardiovascular, neurodegenerative, and genetic diseases. Some have posited that lncRNAs may also have a major role in ADCitation35,Citation38,Citation39 (; ).
Table 1 Dysregulated lncRNAs in Alzheimer’s disease
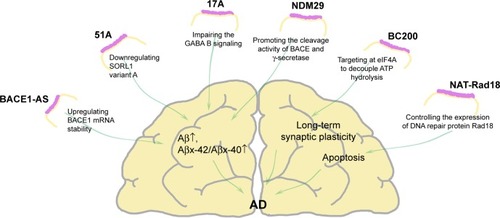
Figure 1 Dysregulated lncRNAs in AD.
Abbreviations: Aβ, amyloid β peptide; AD, Alzheimer’s disease; BACE1, β-site APP cleaving enzyme-1; BC200, brain cytoplasmic 200 RNA; eIF4A, eukaryotic initiation factor 4A; lncRNAs, long noncoding RNAs; NDM29, neuroblastoma differentiation marker 29; SORL1, sortilin-related receptor gene; mRNA, messenger RNA.
BACE1-AS
β-site amyloid precursor protein cleaving enzyme-1 antisense transcript (BACE1-AS) is a conserved RNA transcribed from the positive strand of chromosome 11 on the opposite strand of the BACE1 locus (11q 23.3).Citation16,Citation17 BACE1-AS regulates BACE1 (β-site APP cleaving enzyme-1) expression at both the mRNA and protein levels. BACE1 is essential for the production of the toxic Aβ.Citation40,Citation41 AD pathogenesis has been implicated in many different cell stressors. Following exposure to high temperature, serum starvation, staurosporine, Aβ1–42, high glucose, BACE1-AS, and BACE1 mRNA are both upregulated. This suggests that cell stressors may alter BACE1-AS expression and subsequently BACE1 enzyme activity.Citation16,Citation42 Regardless of whether BACE1-AS is knocked down or overexpressed, both BACE1 mRNA and BACE1 protein are regulated in parallel, thereby reducing Aβ production and plaque deposition.Citation16,Citation17,Citation42 In animals, loss of BACE1 results in numerous behavioral and physiological deficits, including memory loss, reduced synaptic plasticity,Citation43 emotional deficits,Citation44 and peripheral myelination defects.Citation45–Citation49 The delicate physiologic and pathologic boundaries indicate that BACE1 expression should be tightly regulated.Citation16,Citation49
In summary, cell stress increases BACE1-AS levels, which in turn stimulates BACE1 expression, which could enhance APP processing and Aβ1–42 production. Elevated Aβ1–42 levels can further promote BACE1-AS overexpression and the APP processing cascade in a feedforward manner.Citation16,Citation42,Citation50 By forming an RNA duplex, BACE1-AS increases BACE1 mRNA stability.Citation42,Citation51,Citation52 So, BACE1 and BACE1-AS may be potential biomarkers and treatment targets for AD.Citation46,Citation50,Citation53,Citation54
51A
The neuronal sortilin-related receptor gene (SORL1, also known as SORLA and LR11) has long been hypothesized to be involved in AD pathogenesis.Citation55–Citation58 Recent studies have posited that SORL1, as a sorting receptor for APP holoprotein, interacts with APP in endosomes and the trans-Golgi network where it affects trafficking and proteolytic processing.Citation59 Decreased SORL1 expression might shift APP from the retromer recycling pathway to the β-secretase cleavage pathway, increasing secreted APP production and subsequent Aβ formation.Citation59,Citation60 51A is a novel ncRNA that maps in an antisense configuration to intron 1 of the SORL1 gene, the synthesis of which promotes the expression of alternatively spliced SORL1 variants. Notably, 51A is overexpressed in in vitro models and the AD brain. One possible mechanism by which 51A might increase AD susceptibility is by increasing amyloid formation via downregulating SORL1 variant A through alternative splicing.Citation59,Citation61
17A
17A is a 159-nt lncRNA synthesized by RNA polymerase III and maps in intron 3 of G-protein-coupled receptor 51 gene (GPR51); it undergoes alternative splicing, increasing the number of GABA B2 receptor isoforms. GABA B R2 splice variant B may affect GABA B biological function by regulating intracellular 3′–5′-cyclic adenosine monophosphate accumulation and the activation of specific potassium channels. These events would impair GABA B signaling, enhance Aβ secretion, and increase the Aβx-42/Aβx-40 ratio. 17A RNA is upregulated in AD compared with control tissues, suggesting that it could directly or indirectly be involved in the mechanism of AD.Citation52,Citation62,Citation63
NDM29
Neuroblastoma differentiation marker 29 (NDM29) is an RNA polymerase III-transcribed ncRNA. NDM29 synthesis is dose-dependently induced by inflammatory stimulation. The upregulation of NDM29 RNA is accompanied by altered APP modulation. Meanwhile, it can promote the cleavage activities of BACE that, in turn, generates an enhanced amount of APP C-terminal fragments for further processing by the γ-secretase cleavage complex to increase Aβ formation and the Aβx-42/Aβx-40 ratio.Citation12,Citation63,Citation64
Brain cytoplasmic 200 RNA (BC200)
BC200 is a translational regulator that targets eukaryotic initiation factor 4A, thus decoupling adenosine triphosphate hydrolysis from RNA duplex unwinding, modulating local protein synthesis in postsynaptic dendritic microdomains, and contributing to the maintenance of long-term synaptic plasticity.Citation65
One postmortem study found that BC200 RNA levels in cortical areas are reduced by >60% between the ages of 49 and 86 years. Compared with age-matched normal brains, BC200 RNA is significantly upregulated in the AD brain. Moreover, the relative BC200 RNA levels in AD-involved brain areas increase in parallel with disease progression. Still, at least one study reported BC200 downregulation.Citation66 The contradiction between studies may be due to differences in brain regions and varying disease severity, but aberrant BC200 RNA expression in AD is a possibility.Citation67
Relative BC200 RNA levels decrease in dendrites but increase in somata. This divergent expression affects microtubule-dependent transport and could contribute to axonal and dendritic blockage that may be early events in AD. It could eventually contribute to local Aβ generation and subsequent amyloid deposition.Citation24,Citation68 Another group found that BC200 RNA is not affected under apoptotic conditions in vitro and hypothesized that BC200 is only involved in necrosis rather than apoptosis.Citation22
NAT-Rad18
Apoptosis is the main form of programmed cell death, and excessive apoptosis causes progressive cell loss that contributes to many neurodegenerative disorders, including AD. Rad18 is a member of the Rad6 epistasis group, which is responsible for postreplication repair. NAT-Rad18 genes encode for natural antisense transcripts against Rad18, encoding a spectrum of DNA-damaging agents. There is a counterbalanced relationship between Rad18 and NAT-Rad18 in both mRNA and protein level, with Rad18 showing a low expression level. NAT-Rad18 is differentially up-regulated expressed in brain tissues especially cortical neurons following exposure to Aβ. Collectively, this evidence indicates that NAT-Rad18 may be involved in AD via its effects on DNA repair system.Citation69
Conclusion
Almost all lncRNAs related to AD have been listed in this review, but investigation into this field is in the early stage. Since AD was first reported, a century passed before the discovery of basic molecular mechanism. Unquestionably, new information about lncRNAs may light a new beacon in the search for AD treatments. Depending on the mechanism of AD, BACE1-AS, 17A, 51A, and NDM29 directly or indirectly increase Aβ formation and/or the Aβx-42/Aβx-40 ratio. The roles of BC200 and NAT-Rad18 are different. BC200 modulates local protein synthesis to maintain long-term synaptic plasticity. NAT-Rad18 is implicated in apoptosis, while BC200 is only involved in necrosis. As the lncRNA field continues to develop, we still need to elucidate how lncRNAs operate at the molecular and cellular levels. Most recent studies suggest that lncRNAs are desirable candidates in the ongoing quest for AD biomarkers and could help identify rational therapeutic strategies. An enhanced understanding of lncRNA biology could open more avenues to early AD diagnosis and treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
- AnandRGillKDMahdiAATherapeutics of Alzheimer’s disease: past, present and futureNeuropharmacology201476Pt A275023891641
- KalraJKhanAReducing Abeta load and tau phosphorylation: emerging perspective for treating Alzheimer’s diseaseEur J Pharmacol201576457158126209363
- KumarASinghAEkavaliA review on Alzheimer’s disease pathophysiology and its management: an updatePharmacol Rep201567219520325712639
- MullerMKuiperijHBClaassenJAKustersBVerbeekMMMicroRNAs in Alzheimer’s disease: differential expression in hippocam-pus and cell-free cerebrospinal fluidNeurobiol Aging201435115215823962497
- Alzheimer’s Association2015 Alzheimer’s disease facts and figuresAlzheimers Dement201511333238425984581
- ZhangZLong non-coding RNAs in Alzheimer’s diseaseCurr Top Med Chem201616551151926268333
- KangMJAbdelmohsenKHutchisonERHuD regulates coding and noncoding RNA to induce APP-->Abeta processingCell Rep2014751401140924857657
- MattsonMPDegenerative and protective signaling mechanisms in the neurofibrillary pathology of ADNeurobiol Aging1995163447457 discussion 458–4637566352
- HulstaertFBlennowKIvanoiuAImproved discrimination of AD patients using beta-amyloid(1–42) and tau levels in CSFNeurology19995281555156210331678
- TarkowskiELiljerothAMNilssonATNF gene polymorphism and its relation to intracerebral production of TNFalpha and TNFbeta in ADNeurology200054112077208110851366
- AlgarzaeNHebronMMiessauMMoussaCEParkin prevents cortical atrophy and Abeta-induced alterations of brain metabolism: (1)(3)C NMR and magnetic resonance imaging studies in AD modelsNeuroscience2012225223422960314
- MassoneSCiarloEVellaSNDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid beta secretionBiochim Biophys Acta2012182371170117722580042
- Ramirez-BermudezJAlzheimer’s disease: critical notes on the history of a medical conceptArch Med Res201243859559923178566
- YamadaKPatelTKHochgrafeKAnalysis of in vivo turnover of tau in a mouse model of tauopathyMol Neurodegener2015105526502977
- Caillet-BoudinMLBueeLSergeantNLefebvreBRegulation of human MAPT gene expressionMol Neurodegener2015102826170022
- FaghihiMAModarresiFKhalilAMExpression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretaseNat Med200814772373018587408
- ModarresiFFaghihiMAPatelNSSahaganBGWahlestedtCLopez-ToledanoMAKnockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesisInt J Alzheimers Dis2011201192904221785702
- SunXChenWDWangYDbeta-Amyloid: the key peptide in the pathogenesis of Alzheimer’s diseaseFront Pharmacol2015622126483691
- KawabataSHigginsGAGordonJWAmyloid plaques, neurofibrillary tangles and neuronal loss in brains of transgenic mice overexpressing a C-terminal fragment of human amyloid precursor proteinNature199135463534764781793460
- SwerdlowRHPathogenesis of Alzheimer’s diseaseClin Interv Aging20072334735918044185
- TuSOkamotoSLiptonSAXuHOligomeric Abeta-induced synaptic dysfunction in Alzheimer’s diseaseMol Neurodegener201494825394486
- LiuQSunSYuWAltered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cellsJ Neurooncol2015122228329225645334
- Herrera-RiveroMElena Hernandez-AguilarMEmiliano Aranda-AbreuGA strategy focused on MAPT, APP, NCSTN and BACE1 to build blood classifiers for Alzheimer’s diseaseJ Theor Biol2015376323825863267
- ZhouXXuJIdentification of Alzheimer’s disease-associated long noncoding RNAsNeurobiol Aging201536112925293126318290
- MagistriMFaghihiMASt LaurentG3rdWahlestedtCRegulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcriptsTrends Genet201228838939622541732
- KnaussJLSunTRegulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and functionNeuroscience201323520021423337534
- ZhuJFuHWuYZhengXFunction of lncRNAs and approaches to lncRNA-protein interactionsSci China Life Sci2013561087688524091684
- HarrowJFrankishAGonzalezJMGENCODE: the reference human genome annotation for the ENCODE ProjectGenome Res20122291760177422955987
- DerrienTJohnsonRBussottiGThe GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expressionGenome Res20122291775178922955988
- KhorkovaOHsiaoJWahlestedtCBasic biology and therapeutic implications of lncRNAAdv Drug Deliv Rev201587152426024979
- IwakiriJHamadaMAsaiKBioinformatics tools for lncRNA researchBiochim Biophys Acta201618591233026271403
- HamadaMRNA secondary structure prediction from multi-aligned sequencesMethods Mol Biol20151269173825577370
- WilkRHuJBlotskyDKrauseHMDiverse and pervasive subcellular distributions for both coding and long noncoding RNAsGenes Dev201630559460926944682
- WashietlSKellisMGarberMEvolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammalsGenome Res201424461662824429298
- MelissariMTGrotePRoles for long non-coding RNAs in physiology and diseasePflugers Arch Epub201635
- ZengCYuXLaiJYangLChenSLiYOverexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemiaJ Hematol Oncol20158112626545364
- DerrienTGuigoRJohnsonRThe long non-coding RNAs: a new (P) layer in the “Dark Matter”Front Genet2011210722303401
- KrausTFGreinerAGuibourtVLisecKKretzschmarHAIdentification of stably expressed incRNAs as valid endogenous controls for profiling of human gliomaJ Cancer20156211111925561975
- SunMNieFQWangZXDeWInvolvement of incRNA dysregulation in gastric cancerHistol Histopathol2016311333926302456
- MulderSDvan der FlierWMVerheijenJHBACE1 activity in cerebrospinal fluid and its relation to markers of AD pathologyJ Alzheimers Dis201020125326020164582
- DashREmranTBUddinMMIslamAJunaidMMolecular docking of fisetin with AD associated AChE, ABAD and BACE1 proteinsBioinformation201410956256825352723
- LiuTHuangYChenJAttenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1AS expressionMol Med Rep20141031275128124970022
- MaHLesneSKotilinekLInvolvement of beta-site APP cleaving enzyme 1 (BACE1) in amyloid precursor protein-mediated enhancement of memory and activity-dependent synaptic plasticityProc Natl Acad Sci U S A2007104198167817217470798
- LairdFMCaiHSavonenkoAVBACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functionsJ Neurosci20052550116931170916354928
- HuXHicksCWHeWBace1 modulates myelination in the central and peripheral nervous systemNat Neurosci20069121520152517099708
- DecourtBSabbaghMNBACE1 as a potential biomarker for Alzheimer’s diseaseJ Alzheimers Dis201124Suppl 2535921403391
- BorghiRPatriarcaSTraversoNThe increased activity of BACE1 correlates with oxidative stress in Alzheimer’s diseaseNeurobiol Aging20072871009101416769154
- StockleyJHO’NeillCUnderstanding BACE1: essential protease for amyloid-beta production in Alzheimer’s diseaseCell Mol Life Sci200865203265328918695942
- VassarRKandalepasPCThe beta-secretase enzyme BACE1 as a therapeutic target for Alzheimer’s diseaseAlzheimers Res Ther2011332021639952
- DislichBLichtenthalerSFThe membrane-bound aspartyl protease BACE1: molecular and functional properties in Alzheimer’s disease and beyondFront Physiol20123822363289
- YuanJVenkatramanSZhengYMcKeeverBMDillardLWSinghSBStructure-based design of beta-site APP cleaving enzyme 1 (BACE1) inhibitors for the treatment of Alzheimer’s diseaseJ Med Chem201356114156418023509904
- WanPSuWZhuoYThe role of long noncoding RNAs in neurode-generative diseasesMol Neurobiol Epub2016224
- EvinGHinceCBACE1 as a therapeutic target in Alzheimer’s disease: rationale and current statusDrugs Aging2013301075576423842796
- PerneczkyRAlexopoulosPCerebrospinal fluid BACE1 activity and markers of amyloid precursor protein metabolism and axonal degeneration in Alzheimer’s diseaseAlzheimers Dement2014105 SupplS425S429.e42124239250
- JacobsenLMadsenPMoestrupSKMolecular characterization of a novel human hybrid-type receptor that binds the alpha2-macroglobulin receptor-associated proteinJ Biol Chem19962714931379313838940146
- YamazakiHBujoHSaitoYA novel member of the LDL receptor gene family with eleven binding repeats is structurally related to neural adhesion molecules and a yeast vacuolar protein sorting receptorJ Atheroscler Thromb19974120269583350
- LeeJHBarralSReitzCThe neuronal sortilin-related receptor gene SORL1 and late-onset Alzheimer’s diseaseCurr Neurol Neurosci Rep20088538439118713574
- RogaevaEMengYLeeJHThe neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer diseaseNat Genet200739216817717220890
- CiarloEMassoneSPennaIAn intronic ncRNA-dependent regulation of SORL1 expression affecting Abeta formation is upregulated in post-mortem Alzheimer’s disease brain samplesDis Model Mech20136242443322996644
- KhvotchevMSudhofTCProteolytic processing of amyloid-beta precursor protein by secretases does not require cell surface transportJ Biol Chem200427945471014710815316009
- MaQLGalaskoDRRingmanJMReduction of SorLA/LR11, a sorting protein limiting beta-amyloid production, in Alzheimer disease cerebrospinal fluidArch Neurol200966444845719364929
- MassoneSVassalloIFiorinoG17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer diseaseNeurobiol Dis201141230831720888417
- GavazzoPVassalliMCostaDPaganoANovel ncRNAs transcribed by Pol III and elucidation of their functional relevance by biophysical approachesFront Cell Neurosci2013720324223537
- VellaSPennaILongoLPerhexiline maleate enhances antitumor efficacy of cisplatin in neuroblastoma by inducing over-expression of NDM29 ncRNASci Rep201551814426674674
- LinDPestovaTVHellenCUTiedgeHTranslational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanismMol Cell Biol20082893008301918316401
- MusEHofPRTiedgeHDendritic BC200 RNA in aging and in Alzheimer’s diseaseProc Natl Acad Sci U S A200710425106791068417553964
- WuPZuoXDengHLiuXLiuLJiARoles of long noncoding RNAs in brain development, functional diversification and neurode-generative diseasesBrain Res Bull201397698023756188
- IacoangeliABianchiRTiedgeHRegulatory RNAs in brain function and disordersBrain Res20101338364720307503
- ParentiRParatoreSTorrisiACavallaroSA natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosisEur J Neurosci20072692444245717970741
