Abstract
Background
There is a large body of evidence demonstrating long-lasting protective effect of each full-term pregnancy (FTP) on the development of breast cancer (BC) later in life, a phenomenon that could be related to both hormonal and immunological changes during pregnancies. In this work, we studied the pregnancy-associated differences in peripheral blood gene expression profiles between healthy women and women diagnosed with BC in a prospective design.
Methods
Using an integrated system epidemiology approach, we modeled BC incidence as a function of parity in the Norwegian Women and Cancer (NOWAC) cohort (165,000 women) and then tested the resulting mathematical model using gene expression profiles in blood in a nested case–control study (460 invasive case–control pairs) of women from the NOWAC postgenome cohort. Lastly, we undertook a gene set enrichment analysis for immunological gene sets.
Results
A linear trend fitted the dataset precisely showing an 8% decrease in risk of BC for each FTP, independent of stratification on other risk factors and lasting for decades after a woman’s last FTP. Women with six children demonstrated 48% reduction in the incidence of BC compared to nulliparous. When we looked at gene expression, we found that 756 genes showed linear trends in cancer-free controls (false discovery rate [FDR] 5%), but this was not the case for any of the genes in BC cases. Gene set enrichment analysis of immunologic gene sets (C7 collection in Molecular Signatures Database) revealed 215 significantly enriched human gene sets (FDR 5%).
Conclusion
We found marked differences in gene expression and enrichment profiles of immunologic gene sets between BC cases and healthy controls, suggesting an important protective effect of the immune system on BC risk.
Introduction
Each pregnancy represents a unique and remarkable challenge to the mother’s immune system, as her body must accept and carry a fetus to term without immune rejection. Immunological tolerance during pregnancy is necessary for women’s bodies to accept the fetus.Citation1,Citation2 This obligate tolerance has been observed for many years, and because of it, each fetus can be seen as a different semi-allograft. Interestingly, “immune evasion”, which consists of some similar immunological changes, was included as another hallmark of cancer,Citation3 and researchers have recognized some common characteristics between these two highly different biological processes.Citation4–Citation7 Looking at this, it seems paradoxical that increasing number of full-term pregnancies reduces the risk of breast cancer (BC) in women.Citation8,Citation9 Some studies have even shown a protective effect up to eight or nine children.Citation8 However, neither spontaneous nor induced abortions confer the same risk reduction.Citation10,Citation11
The role of hormones is well established in both pregnancy and tumorigenesis; however, the influence of hormonal fluctuations associated with pregnancy on the cancer development in a long-term perspective remains unclear.Citation12,Citation13 High, changing levels of hormones such as estrogens, gestagens, and human chorionic hormone initiate and support immunological tolerance during pregnancy,Citation14 whereas the high levels of estrogens and progestins found in oral contraceptives and hormone replacement therapy are classified as carcinogens.Citation15
We have previously shown that blood gene expression profiles differ between women with BC and BC-free controls, both at time of diagnosisCitation16 and during the years prior to diagnosis.Citation17 Additionally, in a recent study, we compared gene expression in blood and breast tumor tissue.Citation18 We revealed some similarities in expression profiles mainly for immunogenic tumors; however, we demonstrated that there is no obligate common gene expression pattern.
Here, we aim to explore changes in blood gene expression in women with BC and cancer-free controls according to number of full-term pregnancies (FTPs).
Methods
Norwegian Women and Cancer (NOWAC) study
The NOWAC studyCitation19 is a national population-based study in which a random sample of women living in Norway was recruited starting in 1991. Among the 172,748 women invited to participate, 165,227 women replied to at least one questionnaire and make up the study population. Death and emigration status of these women were updated through linkage to the Cause of Death Registry at Statistics Norway and Central Population Registry, respectively. Information on parity was taken from the NOWAC questionnaires. The date of entry into the present study was set as the date of the first questionnaire answered. The exit date from NOWAC was determined either by the date of cancer diagnosis or end of follow-up (2013/12/31). The NOWAC postgenome cohort consisted of 48,692 women randomly sampled from the whole NOWAC study, born in 1943–1957, with a blood sample buffered to preserve the gene expression profile (PAXgene Blood RNA system; Preanalytix/Qiagen, Hilden, Germany). The NOWAC postgenome biobank contained blood samples from women taken by local physicians and mailed overnight to the study center in Tromsø. The blood was collected in 2003–2006.Citation20 BC cases that were diagnosed in 2003–2009 were identified through linkage to the National Cancer Registry of Norway. Altogether 546 cases were identified prospectively, ie, developed BC after blood sample donation.
For each BC case, a control from the NOWAC post-genome cohort was assigned, matched by time of blood sampling and year of birth, to be analyzed together with the case. The controls were used to establish the average (mean) gene expression in individuals without cancer and to serve as the basis for exposure-adjusted analyses. The expression level of a gene not involved in the carcinogenic process varied depending on day-to-day changes in exposures such as environment and nutrition, resulting in random fluctuations of the difference in gene expression between cases and matched controls around a population-average constant over time. On the other hand, the difference in the expression of genes related to different stages of the carcinogenic process varied over time in a nonrandom way, thus exhibiting a nonrandom trend. Moreover, the changes in the expression of genes related to the carcinogenesis could be complicated by other effects caused by exposure to carcinogens.
Laboratory procedures
All extraction and microarray services were provided by the Genomics Core Facility, Norwegian University of Science and Technology, Trondheim, Norway. To control for technical variability such as different batches of reagents and kits, day-to-day variations, microarray production batches, and effects related to different laboratory operators, each case–control pair was kept together throughout all extraction, amplification, and hybridization procedures. RNA extraction was performed using the PAXgene Blood mRNA Isolation kit (Preanalytix/Qiagen) according to the manufacturer’s instructions. RNA quality and purity were assessed using the NanoDrop ND 8000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), respectively. RNA amplification was performed on 96-well plates using 300 ng of total RNA and the Illumina TotalPrep-96 RNA Amplification Kit (Ambio, Inc., Austin, TX, USA). The amplification procedure consisted of reverse transcription with a T7 promoter and ArrayScript, followed by a second-strand synthesis. In vitro transcription with T7 RNA polymerase using a biotin–nucleoside triphosphates mix produced biotinylated cRNA. All case–control pairs were run on either the IlluminaHumanAWG-6 version 3 expression bead chips or the HumanHT-12 version 4 expression bead chips. Outliers were excluded after visual examination of dendrograms, principal component analysis (PCA) plots and density plots. Individuals who were considered borderline outliers were excluded if their laboratory quality measures were below given thresholds (RNA integrity number value <7, 260/280 ratio <2, 260/230 ratio <1.7, and 50 < RNA < 500).
Statistical methods
The first stage of the analysis was to explore the relationship between full-term pregnancies and BC in the whole NOWAC cohort. Based on person-years, the percentage change in incidence rates of BC for each additional child was calculated by using a Poisson linear regression. Each covariate was analyzed separately in a stratified analysis with test for interactions using the Wald test. A Cox proportional semiparametric hazard analysis with attained age as follow-up was used for estimating the overall effect of potential confounders.
Then, gene expression profiles were analyzed for BC cases and controls using microarray technology. The dataset with 30,046 probes for each individual was preprocessed as previously described.Citation21 The dataset was background corrected using negative control probes, log2 transformed using a variance stabilizing technique,Citation22 and quantile normalized. Data from the two Illumina chips (HumanWG-6 v3 and HumanHT-12 v4) were combined on identical nucleotide universal identifiers.Citation23 We retained probes present in at least 70% of the individuals. If a gene was represented with more than one probe, the average expression level of the probes was used as the expression for the gene, resulting in a dataset with 8155 genes. The probes were translated to genes using the lumiHumanIDMapping database.Citation24 Finally, the differences in the log2 gene expression for each case–control pair were computed and used in the statistical analyses. We then excluded 79 case–control pairs in which the case was diagnosed with in situ BC and seven pairs in which the controls were diagnosed with BC before the end of follow-up in 2014. The final preprocessed dataset included 460 case–control pairs.
A nested case–control design was chosen in order to reduce batch effects in the laboratory. In PCA plots, we observed no batch effects in the data that were obtained after we computed differences in log2 gene expression between cases and controls. We used the Bioconductor R-package Limma (linear models for microarrays) to identify the genes that were influenced by parity.Citation25 In the linear model, the responses were the differences in the log2 gene expression for each case–control pair, while we included the parity of the control and the parity of the case as covariates. In the analyses, we merged parities 1–3 and 4–6 so that the parity data consisted of three different values: 0, 1–3, and 4–6. The merging was made in order to reduce the effect of the highest parities. The distribution of parities for cases and controls included in this study is shown in . We identified gene sets that were influenced by parity using Limma in the same way as we did for individual genes by using enrichment scores for gene sets instead of differences in the log2 gene expressions as responses in the linear model. The enrichment scores for gene sets were obtained from the differences in log2 gene expressions using the Bioconductor R-package gene set variation analysis (GSVA).Citation26
Ethics approval and consent to participate
The NOWAC study was approved by the Norwegian Data Inspectorate and the Regional Ethical Committee of North Norway (REK). The study was conducted in compliance with the Declaration of Helsinki, and all participants gave written informed consent. The linkages of the NOWAC database to national registries such as the Cancer Registry of Norway and registries on death and emigration were approved by the Directorate of Health. The women were informed about these linkages. Furthermore, the collection and storing of human biological material was approved by the REK in accordance with the Norwegian Biobank Act. Women were informed in the letter of introduction that the blood samples would be used for gene expression analyses.
Results
Reduction of BC risk after full-term pregnancies
Our first step was to estimate the mathematical relationship between parity and BC incidence in the complete NOWAC cohort, which consisted of 165,227 participants and 6536 registered incident BC cases identified through linkage to the Cancer Registry of Norway. Increasing parity was related to less BC in mothers, less use of hormone replacement therapy, larger body mass index (BMI), earlier first birth, more breast feeding, and less use of oral contraceptives (). Among both premenopausal and postmenopausal women, a regular decrease in incidence rates was found ( and ), and in linear regression analyses of BC incidence in the complete NOWAC cohort, each new pregnancy conferred a strong linear reduction in the absolute risk (8.0%; ). This led to an observed reduction of 48% for a woman with six children.
Figure 1 BC incidence rates in the NOWAC study, 1991–2013.
Abbreviations: BC, breast cancer; CI, confidence interval; NOWAC, Norwegian Women and Cancer; PY, person-years.
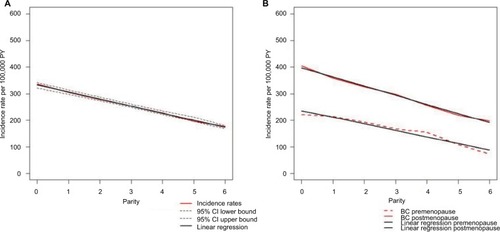
Table 1 Description of the entire study population according to parity
Differences in BC incidence rates were of the same order between nulliparous and uniparous women and for women with five or six children, illustrating an additive effect. The linearity of BC incidence was equally clear regardless of menopausal status, showing the persistent effect of age, but postmenopausal women had higher BC incidence rates (). Stratification on all risk factors (current use of hormone replacement therapy, BMI, age at first birth, breastfeeding, ever use of oral contraceptives, and maternal history of BC) revealed the same linear patterns (). There were no significant interactions between different strata of the risk factors (none in the Wald test). Using the Cox’s multivariate hazard model adjusted for the same risk factors (taken from ), the crude estimated relative risk (RR) in women with six children versus nulliparous women was 0.50 (95% CI 0.37–0.66) and adjusted RR was 0.49 (95% CI 0.36–0.68), similar to the ones derived from Poisson regression analysis ().
Differential pregnancy-associated blood gene expression between cases and controls
This linear relationship independent of other covariates was then used to test the hypothesis that blood gene expression of some genes increased or decreased linearly according to parity.
An overall analysis without parity status showed no significant differences in expressed genes between cases and controls. When we added one parity variable for the cases and one for the controls in the model, we found a significant linear relationship between log gene expression and parity for 756 genes in BC-free controls, but we found no such relationship in BC cases (false discovery rate [FDR] 5%). Among the controls, 96 of 756 significant genes had at least 5% change per child (91 were upregulated and five were downregulated) and four genes had at least 10% change per child (all were upregulated). A description of potential carcinogenic effects in BC of the ten top significant genes showed () that four of them are tumor promoter genes and one is a tumor suppressor gene; however, the function of five significant genes was unknown.
Table 2 Top 10 single genes ranked according to q-value with logFC and their role in BC
Gene set enrichment analysis
demonstrates the results of gene set enrichment analysis. We identified 588 gene sets from the C7 collection in Molecular Signatures Database (MSigDB; http://software.broadinstitute.org/gsea/index.jsp)Citation27 that were significantly enriched when the parity of the controls varied (FDR 5%). Experimentally produced gene sets were submitted to MSigDB from researchers using both in vivo and in vitro materials, as well as both human cells or tissues and animal models. Of our 588 enriched gene sets, 215 were derived from human data and 373 were derived mostly from mouse data.
Table 3 Gene set enrichment analyses for all gene sets included in MSigDB
Detailed characterization of the top 10 gene sets from each species showed a discrepancy in the tissues analyzed: blood and cord blood in humans versus bone marrow and spleen in mice ( and ). We had information on gender for only one of the top 10 human gene sets and four of the mouse gene sets (after contacting main authors of the associated publications). None of the MSigDB gene sets from either humans or mice included information on the number of full-term pregnancies ().
Table 4 Top 10 gene sets for humans and mice with information on experimental design
Discussion
We suggest that our unexpected finding of no changes in gene expression levels from circulating immune-competent cells in BC cases as opposed to parity-dependent changes in healthy women might be linked to the long-term immune-related protection against BC conferred by semi-allograft experience of each pregnancy. Our findings demonstrating the 8% decrease in BC risk for each birth correspond well to a number of previously published studies.Citation8,Citation9
The systems epidemiology approach we used is based on an integrated analysis in the same cohort; first, hypotheses describing the mathematical relationship between parity and BC incidence in the whole NOWAC study are explored. The findings were that the linear relationship between parity and BC could be described as independent of other risk factors. Then, this hypothesis was tested using gene expression profiles obtained from the postgenome biobank of NOWAC. Consequently, we reduced the number of multiple testing in the gene expression analysis and the potential for false-positive findings.
In a recent analysis of interactions between gene expression in BC tissues and blood samples nested in the NOWAC postgenome study,Citation18 we found that genes expressed in tumors and used as markers are not necessarily expressed in a similar manner in blood. Not surprisingly, tumors were enriched for genes related to cancer hallmarks, while in blood, the genes represented either general cellular or immune-related processes.
Currently, there is no unifying concept explaining mechanism of pregnancy-related BC protection. Yet, four main theories may be underlined: decreased number of mammary stem cells, changes in circulating hormone levels, differentiation of breast epithelial cells, and altered estrogen responsiveness.Citation28 Notably, most of these theories are based on studies focusing on the local changes in the mammary gland, while we used blood samples as the source of data, thus looking at systemic processes.
The single-gene analysis revealed a set of genes linearly changing expression after pregnancies in controls. Almost all of these genes were upregulated. This parity-dependent effect was nonsignificant when we compared all cases and controls without taking parity status into account. A possible explanation could be the low parity in the study population with two children as the median value. Of the top ten significant genes, four upregulated genes were known to be related to breast carcinogenesis.Citation29–Citation32 Similar to our results, one study found lower levels of TFRC in plasma of BC cases compared to controls.Citation33 However, the overall meaning of these expression changes in healthy women is uncertain.
Previously, Rotunno et alCitation34 demonstrated the significant enrichment of inflammatory and immune response genes in parous women compared to nulliparous. In our gene set enrichment analysis, we observed significant enrichment of numerous gene sets related to the immune system (C7 collection in MSigDB). It is nearly impossible to make an unambiguous conclusion from these results due to the complexity of gene sets’ data and lack of essential information in gene sets’ description (eg, we were unable to ascertain the gender of blood donors for the majority of human experiments even after contacting the authors of publications). Nevertheless, we observed a clear interspecific difference between components of the immune system: the gene sets were related to (in mice, most of the gene sets were of innate immunity origin, while in humans, they were of adaptive origin) and between the sources of the cells for the experiments. While the latter can be explained by technical and ethical considerations, the former raises yet another concern on the validity of results obtained in animal models.Citation35–Citation39
Pregnancy and cancer share multiple characteristics including proliferation, invasion, neoangiogenesis, and modulation of local and systemic immune response.Citation4 Indeed, in our study, we observed that women with a high parity have a higher expression of tumor promoter genes and a lower expression of tumor suppressor genes; therefore, we propose that the body might experience each pregnancy as a “pseudo cancer”. It is possible to suggest that some pregnancy-related processes may have common genetic signatures and protein expression profile with initial stages of breast tumor formation. Being securely controlled by a variety of local and systemic mechanisms during pregnancy, the immune system does not respond to the changes in breast tissue, while storing memory and effector cells. Later, these cells can initiate rapid and extensive response against cancer cells during the early stages of carcinogenesis. This phenomenon of long-term storage of various subsets of antigen-experienced immune cells after pregnancies has previously been described.Citation40,Citation41 Slight expressional differences in each pregnancy expand the pool of antigen-specific immune cells, thus increasing protective effect with the number of pregnancies. Recently, Krause et alCitation42 published an experimental study confirming our hypothesis.
Our findings of an impaired immune surveillance in BC patients opposed to healthy women may generate novel approaches to immunotherapy based on the natural mechanisms involved in amplified protection against BC with repeated pregnancies. Deciphering of these mechanisms could significantly improve response rate of BC patients to immunotherapy, which at present remains poor.Citation43–Citation45
The major weakness of the study is the lack of statistical power for the gene expression analyses. However, this was compensated by the strict hypothesis testing approach. The strength of the design is the complete follow-up on cancer, emigration, and mortality based on a unique national birth number. The external validity of the parity data in NOWAC was found to be excellent in a linkage study based on information from the fertility register in Statistics Norway.Citation46
Conclusion
To our knowledge, this is the first study revealing the involvement of the immune system in parity-associated BC protection with the use of systems epidemiology approach. Owing to descriptive study design, it is not possible to hypothesize the exact mechanisms responsible for the pregnancy-associated protection from BC using data obtained in this work. Our focus on systemic rather than on local changes may provide the scientific community with a fresh view on the biology of BC and may become a foundation for future research.
Data sharing statement
The datasets are stored at the European Genome-phenome Archive with accession number EGAD00010001062.
Author contributions
EL, JT, and MH carried out conception and design of the study. EL, LH, and MH were involved in the development of methodology. EL, LH, and MH contributed toward acquisition of data. EL, AN, IS, JT, KSO, LH, and MH performed analysis and interpretation of data (eg, statistical analysis, biostatistics, and computational analysis). EL, IS, JT, KSO, LH, and MH carried out writing, review, and/or revision of the manuscript. EL, IS, KSO, LH, and MH provided administrative, technical, or material support. EL conducted study supervision. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Acknowledgments
We are thankful to and impressed by the women who donated blood to the NOWAC postgenome cohort. Bente Augdal, Merete Albertsen, and Knut Hansen were responsible for all infrastructure and administrative issues. The funders had no role in the design of the study; collection, analyses, and interpretation of the data; writing of the manuscript; or decision to submit for publication. This study was supported by a grant from the European Research Council (ERC-AdG 232997 TICE). Some of the data in this article are from the Cancer Registry of Norway. The Cancer Registry of Nor-way is not responsible for the analysis or interpretation of the data presented. Microarray service was provided by the Genomics Core Facility, Norwegian University of Science and Technology, and NMC – a national technology platform supported by the functional genomics program (FUGE) of the Research Council of Norway.
Supplementary materials
Table S1 Distribution of parities for the cases and controls
Table S2 BC cases, PY, and incidence rates per 100,000 PY, according to menopausal status and parity
Table S3 Tests for linear trend between strata of risk factors
Table S4 Age-adjusted and multivariate-adjusted Cox proportional hazard ratios and 95% CIs for BC according to parity in the entire NOWAC cohort
Table S5 Full names of gene sets described in
Disclosure
The authors report no conflicts of interest in this work.
References
- ErlebacherAMechanisms of T cell tolerance towards the allogeneic fetusNat Rev Immunol2012131233323237963
- WilliamsZInducing tolerance to pregnancyN Engl J Med2012367121159116122992082
- HanahanDWeinbergRAHallmarks of cancer: the next generationCell2011144564667421376230
- HoltanSGCreedonDJHaluskaPMarkovicSNCancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agentsMayo Clin Proc20098411985100019880689
- CuriglianoGImmunity and autoimmunity: revising the concepts of response to breast cancerBreast201120S71S7422015297
- ElinavENowarskiRThaissCAHuBJinCFlavellRAInflammation-induced cancer: crosstalk between tumours, immune cells and microorganismsNat Rev Cancer2013131175977124154716
- ZhaoHOzenMWongRJStevensonDKHeme oxygenase-1 in pregnancy and cancer: similarities in cellular invasion, cytoprotection, angiogenesis, and immunomodulationFront Pharmacol2014529525642189
- LundEChildbearing in marriage and mortality from breast cancer in NorwayInt J Epidemiol19901935275312262244
- Collaborative Group on Hormonal Factors in Breast CancerBreast cancer and breastfeeding: collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the diseaseLancet2002360932818719512133652
- ReevesGKKanSWKeyTBreast cancer risk in relation to abortion: results from the EPIC studyInt J Cancer200611971741174516646050
- GuoJHuangYYangLAssociation between abortion and breast cancer: an updated systematic review and meta-analysis based on prospective studiesCancer Causes Control201526681181925779378
- TalRTaylorHSBurneyROMooneySBGiudiceLCEndocrinology of PregnancySouth Dartmouth, MAMDText.com2000
- HendersonBEPonderBAJRossRKHormones, Genes, and CancerOxford New YorkOxford University Press2003
- RobinsonDPKleinSLPregnancy and pregnancy-associated hormones alter immune responses and disease pathogenesisHorm Behav201262326327122406114
- IARCHormonal Contraception and Post-menopausal Hormonal TherapyIARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 72LyonIARC1999
- LundEHoldenLBøvelstadHA new statistical method for curve group analysis of longitudinal gene expression data illustrated for breast cancer in the NOWAC postgenome cohort as a proof of principleBMC Med Res Methodol20161612826944545
- DumeauxVUrsini-SiegelJFlatbergAPeripheral blood cells inform on the presence of breast cancer: a population-based case-control studyInt J Cancer2015136365666724931809
- DumeauxVFjukstadBFjosneHEInteractions between the tumor and the blood systemic response of breast cancer patientsPLoS Comput Biol2017139e100568028957325
- LundEDumeauxVBraatenTCohort profile: the Norwegian women and cancer study – NOWAC – Kvinner og kreftInt J Epidemiol2007371364117644530
- DumeauxVBørresen-DaleA-LFrantzenJ-OKumleMKristensenVNLundEGene expression analyses in breast cancer epidemiology: the Norwegian women and cancer postgenome cohort studyBreast Cancer Res2008101R1318271962
- HoldenMHoldenLStatistical Analysis of Gene Expression in Blood Before Diagnosis of Breast CancerNR note, SAMBA/07/162016 Available from: https://www.nr.no/files/samba/smbi/note2016SAMBA0716BreastCancer.pdfAccessed July 5, 2018
- LinSMDuPHuberWKibbeWAModel-based variance-stabilizing transformation for Illumina microarray dataNucleic Acids Res2008362e1118178591
- DuPKibbeWALinSMnuID: a universal naming schema of oligonucleotides for Illumina, Affymetrix, and other microarraysBiol Direct2007211617540033
- DuPFengGKibbeWLinSlumiHumanIDMapping: Illumina Identifier mapping for Human2016R package version 1.10.1 Available from: https://bioconductor.org/packages/release/data/annotation/html/lumiHumanIDMapping.htmlAccessed July 5, 2018
- SmythGKLinear models and empirical Bayes methods for assessing differential expression in microarray experimentsStat Appl Genet Mol Biol200431125
- HänzelmannSCasteloRGuinneyJGSVA: gene set variation analysis for microarray and RNA-Seq dataBMC Bioinformatics2013141723323831
- SubramanianATamayoPMoothaVKGene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profilesProc Natl Acad Sci USA200510243155451555016199517
- BrittKAshworthASmalleyMPregnancy and the risk of breast cancerEndocr Relat Cancer200714490793318045947
- AslamAMittalSKochFAndrauJCWinklerGSThe Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferationMol Biol Cell200920173840385019605561
- YangYHMarkusMAMangsAHRaitskinOSperlingRMorrisBJZRANB2 localizes to supraspliceosomes and influences the alternative splicing of multiple genes in the transcriptomeMol Biol Rep20134095381539523666063
- DanielsTRDelgadoTRodriguezJAHelgueraGPenichetMLThe transferrin receptor part I: biology and targeting with cytotoxic antibodies for the treatment of cancerClin Immunol2006121214415816904380
- ZhangHLuHXiangLHIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cellsProc Natl Acad Sci U S A201511245E6215E622326512116
- BuasMFRhoJHChaiXZhangYLampePDLiCICandidate early detection protein biomarkers for ER+/PR+ invasive ductal breast carcinoma identified using pre-clinical plasma from the WHI observational studyBreast Cancer Res Treat2015153244545426319120
- RotunnoMSunXFigueroaJParity-related molecular signatures and breast cancer subtypes by estrogen receptor statusBreast Cancer Res2014164R7425005139
- BeuraLKHamiltonSEBiKNormalizing the environment recapitulates adult human immune traits in laboratory miceNature2016532760051251627096360
- AnisimovVNUkraintsevaSVYashinAICancer in rodents: does it tell us about cancer in humans?Nat Rev Cancer200551080781916195752
- BeckersJWurstWde AngelisMHTowards better mouse models: enhanced genotypes, systemic phenotyping and envirotype modellingNat Rev Genet200910637138019434078
- No authors listedOf men, not miceNat Med201319437923558605
- MakIWEvaniewNGhertMLost in translation: animal models and clinical trials in cancer treatmentAm J Transl Res20146211411824489990
- RoweJHErteltJMXinLWaySSPregnancy imprints regulatory memory that sustains anergy to fetal antigenNature2012490741810210623023128
- MorGAldoPAlveroABThe unique immunological and microbial aspects of pregnancyNat Rev Immunol201717846948228627518
- KrauseALSchuetzFBoudewijnsMParity improves anti-tumor immunity in breast cancer patientsOncotarget201786210498110499129285226
- VonderheideRHDomchekSMClarkASImmunotherapy for breast cancer: what are we missing?Clin Cancer Res201723112640264628572258
- SalatinoMGirottiMRRabinovichGAGlycans pave the way for immunotherapy in triple-negative breast cancerCancer Cell201833215515729438689
- TolbaMFOmarHAImmunotherapy, an evolving approach for the management of triple negative breast cancer: converting non-responders to respondersCrit Rev Oncol Hematol201812220220729373180
- LundEKumleMBraatenTExternal validity in a population-based national prospective study – the Norwegian Women and Cancer Study (NOWAC)Cancer Causes Control200314101001100814750540
