Abstract
Purpose
This research aimed to develop and validate a META-algorithm combining individual immune-mediated inflammatory disease (IMID)-specific algorithms to identify the exact IMID indications for incident biological drug users from claims data within the context of the Italian VALORE project.
Methods and Patients
All subjects with at least one dispensing of TNF-alpha inhibitors, anti-interleukin agents, and selective immunosuppressants approved for IMIDs were identified from claims databases of Latium region in Italy (observation period: 2010–2020). Validated coding algorithms for identifying individual IMIDs from claims databases were found from published literature and combined into a META-algorithm. Positive predictive value (PPV), sensitivity (Se), negative predictive value (NPV), specificity (Sp), and accuracy (Acc) were estimated for each indication against the electronic therapeutic plans (ETPs) of the Latium region as the reference standard. Lastly, the frequency of the indication of use across individual biologic drugs was compared with that reported in three other Italian regions (Lombardy, Apulia, and the Veneto region).
Results
In total, 9755 incident biological drug users with a single IMID indication were identified. Using the newly developed META-algorithm, an indication of use was detected in 95% (n=9255) of the total cohort. The estimated Acc, Se, Sp, PPV, and NPV, against the reference standard were as follows: 0.96, 0.86, 0.97, 0.82, and 0.98 for Crohn’s disease, 0.96, 0.80, 0.98, 0.85, and 0.97 for ulcerative colitis, 0.93, 0.76, 0.99, 0.95, and 0.92 for rheumatoid arthritis, 0.97, 0.75, 0.99, 0.85, and 0.98 for spondylarthritis, and 0.91, 0.92, 0.91, 0.88, and 0.94 for psoriatic arthritis/psoriasis, respectively. Additionally, no substantial difference was observed in the frequency of indication of use by active ingredient among Latium and the other three Italian regions included in the study.
Conclusion
The newly developed META-algorithm demonstrated high validity estimates in the Italian claims data and was capable of discriminating with good performance among the most frequent IMID indications.
Plain Language Summary
In the claims database, the lack of information on the indication of use represents a well-known limitation for the conduct of observational studies. This study was conducted to develop and validate a META-algorithm that accurately identifies the exact indication for the use of biological drugs in treating various immune-mediated inflammatory diseases. Using claims databases from the Latium region, we developed and validated a META-algorithm. The META-algorithm combines disease-specific algorithms for different immune-mediated inflammatory diseases (ie, Crohn’s disease, ulcerative colitis, rheumatoid arthritis, spondyloarthritis, psoriasis, and psoriatic arthritis) and was tested against a reference standard (electronic therapeutic plans of the Lazio region). The META-algorithm reported high validity estimates and was able to distinguish with a good performance among the most frequent IMIDs as indications for use. Applying this META-algorithm may facilitate post-marketing surveillance of biological drugs such as TNF-alpha inhibitors, anti-interleukin, and selective immunosuppressants in specific therapeutic areas in an Italian setting.
Introduction
Since the late 1990s, biological drugs such as Tumor Necrosis Factor (TNF)-alpha inhibitors – including infliximab, adalimumab, etanercept, golimumab, and certolizumab – have been introduced to the market, thus changing dramatically the treatment landscape for various rheumatic, dermatological, and gastrointestinal immune-mediated inflammatory diseases (IMIDs).Citation1 More recently, also other biological drugs as selective immunosuppressant biological drugs (abatacept), anti-integrin (vedolizumab) and anti-interleukins (eg, tocilizumab, secukinumab, and ustekinumab) were approved for such diseases. IMIDs have a prevalence of 5–7% in the general population and include a heterogeneous group of conditions caused by deregulation of the body’s cytokine environment.Citation2 The most common IMIDs for which TNF-alpha inhibitors, anti-interleukin, and selective immunosuppressants are approved are rheumatoid arthritis (RA), spondylarthritis (SpA), psoriasis (PsO), psoriatic arthritis (PsA) and inflammatory bowel diseases (Crohn’s disease - CD and ulcerative colitis – CU). Other IMIDs with lower frequency are hidradenitis suppurativa (HS) and uveitis.Citation3,Citation4 According to the national and international guidelines, biological drugs approved for IMID usually represent the 2nd line treatment in patients who have failed or with contraindications to conventional disease-modifying antirheumatic drugs (DMARDs). Given the recent marketing of biosimilars on a side and, on the other side, the need to monitor (long-term) safety and effectiveness of all biological drugs, intensive post-marketing surveillance of biological drugs in the real-world setting is of paramount importance.
With respect to that, claims databases may be used as source for real-world evidence generation at population-based level and, as such, have been increasingly employed worldwide to provide information on effectiveness and safety of drugs including biological originator and biosimilar drugs in real-world setting.Citation5–8 Such databases can be used to identify both acute and chronic diseases through specific case-identification algorithms that combine information coded in different claims databanks such as hospitalizations and exemptions from co-payment.Citation9 However, in pharmacoepidemiology, it is essential to validate the newly developed algorithms for the identification of outcomes, covariates, as well as indication of use.Citation9–12 In claims database the lack of information on the indication of use represents a well-known limitation for the conduct of observational studies. Understanding the specific reason why a medication was prescribed is crucial for accurately interpreting outcomes. Without this information, researchers may face challenges in properly adjusting for confounding variables and attributing observed effects to the treatment under investigation. Specifically, this holds true for biological drugs such as TNF-alpha inhibitors, anti-interleukin, and selective immunosuppressants as most of these drugs are approved for the treatment of several IMIDs and IMID patients can suffer multiple IMIDs simultaneously,Citation13–15 thus making challenging the identification of the exact indication for the dispensed biological drugs. Despite several algorithms have been previously developed and validated to identify the most frequent rheumatic, dermatological and gastrointestinal IMID individually in claims databases,Citation16–19 coding algorithm for the specific identification of the exact indication for biological drug users using claims databases, has not been developed so far. The aim of this study was to newly develop and validate a META-algorithm for the identification of exact indication of use for biological drugs approved for IMIDs by combining all single disease-specific algorithms for single IMID identification.Citation8
Materials and Methods
Data Source
A validation study was conducted using data extracted from the claims database of the Latium region, which collects information about 5.9 million inhabitants (study period 2010–2020). The Latium region claims database is composed by several databanks such as: (1) inhabitant registry, including demographic information about the year of birth, sex, and date of registration in the regional healthcare system; (2) pharmacy claims database, which collects information about the anatomical therapeutic chemical (ATC) classification system and the Italian marketing authorization code of drug dispensing that are reimbursed by the Italian national health system, such as the biological drugs; (3) hospital discharge records; (4) exemptions from healthcare service co-payment; and (5) outpatients encounter data. Moreover, the Latium region also collects electronic therapeutic plans (ETPs) of patients that are treated with biological drugs, containing very useful clinical information including the exact indication of use. These plans are not digitally available in most of Italian regions where a specialist physician fills them only as paper forms. Finally, in Latium vs other three Italian regions, all coming from the VALORE project network databases, the frequency of the indication for use for the incident biological drug users, as identified by the META-algorithm, was measured and compared. The VALORE project distributed database network was described elsewhere.Citation8,Citation20
Study Population
Biological drug users (approved for at least one IMID under study) with at least one year of look-back period and one year of follow-up were selected from the Latium claims data. Only incident patients with a first dispensing during study period were included in the study (no dispensing of biological drug in the previous years). The date of the first dispensing of the biological drug was considered as the index date. Among incident biological users, patients with only one IMID indication reported in the ETP within one-year pre- and one-year post-index date were included. The following users of TNF-alpha inhibitors, anti-interleukin, and selective immunosuppressant drugs approved for IMID treatment were included in the study: a) TNF-alpha inhibitors: adalimumab (L04AB04), certolizumab pegol (L04AB05), etanercept (L04AB01), golimumab (L04AB06) and infliximab (L04AB02); b) Interleukin inhibitors: anakinra (L04AC03), brodalumab (L04AC12), guselkumab (L04AC16), ixekizumab (L04AC13), sarilumab (L04AC14), secukinumab (L04AC10), tocilizumab (L04AC07), and ustekinumab (L04AC05); c) selective immunosuppressive agent: abatacept (L04AA24); d) anti-integrin: vedolizumab (L04AA33). Rituximab was not included in the study as this biological drug is mostly used in the hematological setting.Citation21,Citation22
Reference Standard
The ETP records for biological drug users approved for IMID treatment were considered as the reference standard. From this registry, the following indications of use were extracted: RA, SpA, PsO, PsA, CD, CU, HS, and uveitis. This databank can be linked to the other databanks by a unique anonymized patient identifier, thus allowing the measurement of the validity estimates of the newly developed META-algorithm using the information from the other available claims data.
Identification of Coding Algorithms for Single IMIDs from the Literature
Based on published articles, coding algorithms to ascertain IMIDs (RA, SpA, PsO, PsA, CD, CU, HS, and uveitis) from claims Italian databases were identified.Citation19,Citation23–31 It was decided to consider only algorithms validated in Italy as those developed using claims databases from other countries may not be directly applicable to the Italian claims databases.Citation9 If no coding algorithms were previously validated in the Italian claims databases, then algorithms developed in other countries were considered. In the algorithms for each IMID indication are reported (ATC and national drug codes used in the indication for use-specific algorithm are described in the appendix – Table S1).
Table 1 Individual IMID-Specific Algorithms Retrieved from the Literature
META-Algorithm Validation
The disease-specific algorithms retrieved from literature were combined into a META-algorithm which was applied to all available look-back and follow-up. The closest diagnosis to the index date (pre- or post-) identified with the META-algorithm was taken as the indication to be validated. In case the META-algorithm found more than one indication for the same patient, two sensitivity analyses were conducted: 1) the priority was given to the indication identified by an exemption from the co-payment record; 2) the priority was given to the indication with the highest number of matches. The number of matches is defined as the number of times an individual user is identified with a specified indication by the META-algorithm during all available look-back and follow-up period. (See Figure S1)
The following validity estimates were calculated against the reference standard to measure the performance of the META-algorithm for the identification of each indication: sensitivity (Se), specificity (Sp), positive predictive value (PPV), negative predictive value (NPV), and accuracy (Acc). In addition, the Youden index and the F-score were calculated. Moreover, the indications retrieved for the false negative patients (those subjects without or with a wrong indication identified from META-algorithm) were described. According to these results, the final version of the META-algorithm was chosen.
The META-algorithm searches for the indication of use in all available look-back and follow-up periods; however, it is unknown whether and to which extent look-back and follow-up may have an impact in identifying the indication of use. As such, the effect of the length of look-back and follow-up on the validity estimates was evaluated by stratifying the cohort according to the calendar year corresponding to the beginning of the treatment with the index drug (three periods with the same length): 2011–2013 (short look-back but long follow-up period), 2014–2016 (long look-back and follow-up period), and 2017–2019 (long look-back but short follow-up period).
Finally, contour plots were generated to represent how the length of follow-up and look-back period influenced the SE, Sp and missingness of the cohort 2014–2016. This cohort was chosen because we hypothesized to have a similar length of look-back and follow-up period.
Application of the META-Algorithm to the VALORE Project Database Network
The validated META-algorithm was then employed to measure the frequency of the indications for the use of incident users of biological drugs in the Latium region. These frequencies were compared to those obtained from the Lombardy, Veneto, and Puglia regions, all of which are part of the VALORE project distributed database network, as an indirect measure of the generalizability of the META algorithm. All statistical analyses were carried out using the R software environment (ver.4.3.0). Data manipulation and visualization were performed using “dplyr” and “ggplot2” packages, respectively.
Results
Starting from a total population of around 5 million inhabitants in the Latium region, 32,405 (0.6%) prevalent users of biological drugs were identified during the study period. Of them, 9755 (30.1%) incident users with only one ETP reporting IMID indication and with at least one year of look-back and one year of follow-up were included in the study ().
Figure 1 Flow chart of incident biological drug users included in the study.

META-Algorithm Validation
Retrieved individual IMID algorithms were combined into a single META-algorithm which was applied to the study cohort of the Latium region: overall, the algorithm was able to attribute the indication in 9255/9755 incident biological users (94.9%): in 2685 (27.5%) of them, the indication was PsO, followed by 2022 (20.7%) for RA, 1652 for PsA (16.9%), 1193 (12.2%) for CD, 1042 (10.6%) for UC, 631 for SpA (6.4%), 28 (0.02%) for HS and in only 2 for uveitis.
The META-algorithm was then tested against the reference standard (ETPs). reports the overall validity estimates of the META-algorithm for the main analysis (closest indication to the index biological drugs). The META-algorithm yielded high (>0.80) and very high (>0.90) estimates of Acc and Sp for each IMID. As for PPV and Se, the META-algorithm reported low estimates values only for PsO (PPV: 0.48), PsA (Se: 0.48), HS (Se: 0.33), and uveitis (Se: 0.13). As for CD, the Youden index was 0.84, while for CU, PsO, AR, and SpA ranged from 0.72 to 0.78. Values of the Youden index lower than 0.60 were found for PsA (0.43), HS (0.33), and uveitis (0.13). The highest F-score was found for CD (0.84), while the lowest was for HS (0.5) and Uveitis (0.23).
Table 2 Validity Estimates of the META-Algorithm According to the Main Analysis
The results of the sensitivity analyses are presented in the Supplementary Materials. In the case of more than one retrieved indication, it was primarily considered the indication identified using exemption from co-payment codes (first sensitivity analysis: Table S2) or the one with the highest number of matches (second sensitivity analysis: Table S3). As for the first sensitivity analysis, results were in contrast with the primary analysis for some IMIDs: ie, for CD and UC estimates values were lower in the sensitivity than in the main analysis (Se of CD: from 0.86 to 0.74; PPV of UC: from 0.85 to 0.74), while greater performance was reported when considering some estimates for RA and PsA (Se of RA: from 0.76 to 0.86; Se of PsA: from 0.48 to 0.66). Instead, PsO reported a lower Se (from 0.89 to 0.75) in sensitivity vs the main analysis but an increase in PPV (0.48 vs 0.61). PsA indication reported for both the Youden index and F-score substantially higher estimates with respect to the main analysis (Youden index: from 0.43 to 0.60; F-score: from 0.59 to 0.72). As for the second sensitivity analysis, only in a few cases the validity estimates were slightly higher than the main analysis (eg, Se of CD: 0.86 vs 0.88; Se of PsO: 0.89 vs 0.93), while a dramatic drop for other validity estimates (eg, PPV of CD: 0.82 vs 0.72; Se of CU: 0.80 vs 0.62; Se of PsA 0.48 vs 0.24) was observed. No indication of use reported substantially higher Youden Index and F-score with respect to the main analysis.
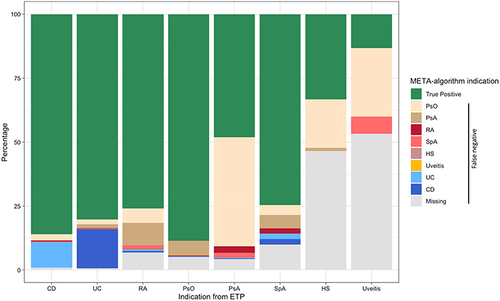
shows that the most common misclassification for CD is UC (10.0%) and for UC is CD (15.3%). Most of PsA patients were misclassified as PsO (42.6%), while only 5.8% of patients with PsO were misclassified as PsA. Finally, the percentage of missingness of the indication of use was lower than 10% for CD (0.9%), UC (0.6%), RA (6.9%), PsO (5.1%), and PsA (4.3%) and SpA (9.9%) indications, while was much higher for HS (46.5%) and uveitis (53.3%), as indications.
Figure 2 Percentage of indications of use assigned by the META-algorithm against the reference standard.
According to these findings, the META-algorithm was modified by combining the PsO and PsA algorithms into a unique indication and retaining the closest indication to the index date as the most accurate method since the first sensitivity analysis mainly improved the values of PsA for which the algorithm was modified. Moreover, HS and Uveitis could not be tracked correctly. The META-algorithm was then launched against the reference standard again with such modification. As for the combined PsO-PsA indication, the META-algorithm reported the following validity estimates: Acc: 0.91, Se: 0.92; Sp: 0.91; PPV: 0.88; NPV: 0.94; Youden: 0.83, F-score: 0.90. Given the high values of the validity estimates for the combined algorithm of PsO and PsA, this version of the META-algorithm was considered as the final version.
Influence of Calendar Time on the Validity Estimates
The cohort stratification according to the index date showed that the estimates could differ based on follow-up and look-back time available. The median look-back and follow-up time for the three cohorts is reported in Figure S2. Table S4 shows that some validity estimates regarding CD and SpA differ among the cohorts: the PPV of CD changed from 0.87 (cohort 2011–2013) to 0.77 (cohort 2017–2019), while the Se of SpA changed from 0.80 (cohort 2011–2013) to 0.69 (cohort 2017–2019). Figure S3 shows how the length of follow-up and length of look-back period may influence Se, Sp, and Acc in the cohort 2014–2016. For each indication, each combination of length of follow-up and look-back was found with levels of Sp and Acc at least over 0.70. At the same time, Se changed dramatically according to the length of the timeframe. As for Se, the slope of level curves was more vertically oriented in favor of the look-back period. This was particularly evident for all indications except for the SpA. Finally, Figure S4 represents how the length of look-back and follow-up influenced the percentage of missing indications. Also, in this case, the slope of level curves was more vertically oriented in favor of the look-back period for the identification of at least one indication of use. For instance, having 24 months of look-back and 12 of follow-up ensures to find an indication of use in at least 67% of cases. Conversely, with a 12-month look-back period and 24 months of follow-up, the percentage of identified indications reaches at least 59% of cases.
Application of the META-Algorithm to Other Regions from the VALORE Project Network
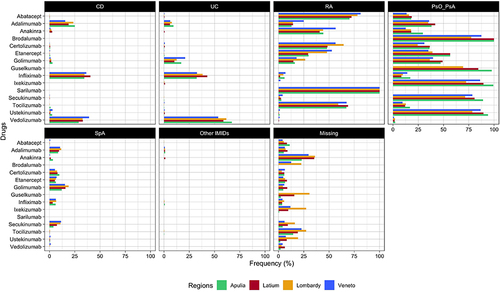
Finally, the frequency of the indications for use of incident users of biologics was measured and compared in Latium versus other three Italian regions (Veneto, Lombardy, and Apulia) by individual compound to assess the external validity of the META-algorithm (). In addition to the patients from Latium, 17,201, 12,044, and 13,776 incident users of biological drugs (2010–2020) and with at least one year of follow-up and one year of the look-back period were identified from Lombardy, Veneto, and Apulia regions, respectively. Overall, no substantial difference in the frequency of indications of biological drug users across regions was observed. Some biological drugs such as anakinra (Latium 35.3%, Apulia: 23%, Lombardy: 36.2%, Veneto: 29.6%) and tocilizumab (Latium: 19.0%, Apulia: 14.5%, Lombardy: 27.3%, Veneto: 22.7%) had higher frequency of missing indication, as compared to other biological drugs for which the frequency of missing indication was under the 10% in all the regions. Instead, ixekizumab and guselkumab had a higher frequency of missingness in the Lombardy region (27% and 30%, respectively) compared to the other regions. Finally, the frequency of misclassified indications (eg, certolizumab for UC or CD) was observed only in a small percentage of patients included (<5%).
Figure 3 Frequency (%) of indication of use stratified by single biological drug across regions (Apulia, Latium, Lombardy, and Veneto).
Discussion
This study provides valuable evidence for researchers to investigate the real-world outcomes and utilization of biological drugs approved for IMIDs, enabling the identification of indications of use from Italian claims data. Other validation studies,Citation19,Citation23,Citation24,Citation32 which were considered for the development of the META-algorithm, were aimed to develop and validate a coding algorithm for the identification of a single IMID in the general population; however, to the best of our knowledge, no previous studies developed and validated a coding algorithm, using claims data, to correctly identify the correct indication of use for biological drug users (eg TNF-alpha inhibitors, anti-interleukin, and selective immunosuppressants), which are approved for IMID treatment. As indication of use is typically not specifically reported in the claims database, the findings of this study can help the conduction of future observational studies on biological drugs in IMID patients by identifying the correct indication of use. Those biological drugs are approved for multiple IMIDs which can also co-occur and given that baseline risk of various safety outcomes may vary substantially across indications of use of biological users the correct identification of the indication of use of biological drugs approved for IMID is of paramount importance for instance for studies assessing comparative safety profiles of those drugs in a real-world setting. Overall, the newly developed META-algorithm identified CD, UC, RA, and SpA as indications for use of biological drugs with high/very high estimate values (PPV, Se, NPV, and Sp). This study demonstrated that the indication of PsA from claims data in almost half of the cases was misclassified as PsO and, as such, PsO and PsA were finally combined as a unique indication into the META-algorithm. PsA often occurs after a PsO diagnosis (only 15% of patients developed PsA without having a pre-existing condition of PsOCitation33). Consequently, the distinction of these two indications using claims data is not accurate. In addition, this study also demonstrated that uveitis and HS could not be tracked with high Acc using claims data. The challenge in identifying uveitis/HS in claims data may be due to a) the low severity of these diseases as compared to other IMID, which may lead to a lower risk of hospitalization or access to healthcare facilities, thus limiting tracing of these diseases using claims data; or b) to the low Se of the disease-specific algorithms included in the META-algorithm which were validated in a non-Italian setting. According to the first hypothesis, inflammatory bowel diseases (CD and UC), which can lead patients more often to access healthcare facilities due to surgery/hospitalizations and require more frequent hospital care, had the lowest levels of missingness among IMIDs (0.9% and 0.6%, respectively). This can be particularly true for the Italian setting (universal, single-payer healthcare system), where claims data are derived from the reimbursement of healthcare services. Moreover, it is necessary to highlight that patients with a single EPT record of Uveitis (n=15) and HS (n=84) were a small number compared to the other IMID indications, which could have impaired the precision of the validity estimates obtained. Nevertheless, as for the most common IMIDs indicated for TNF-alpha inhibitors, anti-interleukin, and selective immunosuppressants (ie, UC, CD, RA, PsO-PsA, and SpA), the META-algorithm reported high (≥0.80) or very high (≥0.90) estimates values with a missing indication lower than 10%.
The comparison between the main analysis (closest indication to index date) and the first sensitivity analysis (priority to an exemption from co-payments) highlighted a difference in the calculated estimates among different single indications of use: ie, Youden Index and/or F-score were higher for PsA, and RA, lower for PsO and inflammatory bowel diseases. The explanation for the observed difference between the main and the first sensitivity analysis relies on the timing and frequency of occurrence of single IMIDs. For instance, PsA occurs in 85% of cases after a PsO diagnosis, so the closest match can likely be a match of a co-existing PsO disease. On the other hand, for other diseases such as CD and UC as well as PsO, by choosing the closest indication, we can prevent the identification of other concomitant IMIDs being diagnosed during follow-up, thus improving validity estimates. As for the second sensitivity analysis, choosing the IMIDs with the highest number of matches did not substantially improve the validity estimates. This can be explained by two reasons: 1) the identification of the indication with the highest number of matches is highly influenced by the number of variables used for the definitions of the single indication-specific algorithms which may lead to the incorrect indication attribution; 2) by choosing IMIDs with the higher number of matches during all available look-back and follow-up the META-algorithm can identify indication of use which are not close the index dispensation of biologic drug thus lowering the validity estimates.
This study also reports that the available length of follow-up and look-back might influence the validity estimates of the META-algorithm. In the cohort restricted to the calendar years 2017–2019 (long look-back but short follow-up period), a drop (>0.10) of the PPV of CD and a drop of the Se of SpA was observed. Notably, clinical guidelines recommend starting biological drug treatment only in IMID patients with a moderate-to-severe IMID:Citation34,Citation35 Since the META algorithm seeks the nearest indication to the index date, irrespective of the identification of a related code before or after the index date, we hypothesize that during the follow-up period, patients may engage more frequently with healthcare services due to potential disease complications. Nevertheless, in patients who have the same length of follow-up and look-back period (ie, cohort 2014–2016), the identification of the indication occurs earlier in the look-back than in the follow-up period.
Overall, we did not observe any substantial difference in the distribution of the frequency of indications for use between Latium (central Italy) and three other Italian regions, including two from northern Italy (Lombardy and Veneto) and one from southern Italy (Apulia). According to this result, the validity of the META-algorithm can also be generalized to other Italian regions. Overall, the frequency of the missingness of the indication of use per single drug was generally lower than 10–15% except for anakinra and tocilizumab, for which the missing indication was higher than this threshold in each region included in the study. This can be related to other non-IMID indications for which these drugs are approved: excluding COVID-19 (our cohort ended in 2020, and the use of these drugs in the first year of the pandemic can be limited), anakinra, which had the higher percentage of missing indication is also approved for periodic fever syndromes as well as familial Mediterranean fever and cryopyrin-associated periodic syndromes. Finally, the META-algorithm reported a good performance in distinguishing the indications of use for CD and UC: this can be observed for golimumab, which is approved for UC but not for CD ().
Strengths and Limitations
This study has several strengths: first, to validate the META-algorithm, a large cohort of biological drug users was included (about 10,000 patients for the validation analysis and about 50,000 to measure the frequency of the indication of use of biological drug users across the regions). Second, several sensitivity analyses were conducted to explore the robustness of the study findings. Finally, indirect methods have been employed to confirm the generalizability of the study findings to other Italian regions. Moreover, since the META-algorithm developed in this study was based mainly on ICD-9-CM and ATC codes, which are globally used to code for diagnoses and drugs, the proposed algorithm is expected to be adaptable to healthcare databases from other countries.
However, some limitations must be acknowledged. First, the META-algorithm is not capable to identify the indication of use for the entire cohort of incident users of biological drugs (about 5% had a missing indication): this is not a limit of the META-algorithm itself but is due to the nature of claims data as previously discussed. Second, the META-algorithm may misclassify some indications of use for biological drugs (eg, certolizumab utilization in patients with inflammatory bowel diseases), although the impact of such misclassification should be limited since its validity estimates are found to be high. Third, the RA algorithm also includes codes of Juvenile Idiopathic Arthritis and Still disease, which were thus not separately identified. However, a very low prevalence of these diseases is reported in the general population, which is unlikely to affect study estimates. Finally, the META-algorithm did not consider censoring biological treatment discontinuation during follow-up and the match with an IMID indication of use could also be found after treatment interruption of the biological drug. To carefully evaluate the impact of drug discontinuation on the calculated validity estimates of indication of use of biological drugs, we conducted a sensitivity analysis by censoring patients at drug discontinuation (60 days of grace period after the end of drug coverage) or switching to another biological drug, which yielded similar results as compared to the main analysis (Table S5).
Conclusions
In conclusion, a META-algorithm for identifying the indication of use for biological drug users from claims data was newly developed and validated in the Italian setting. Our findings reveal high validity estimates for this META-algorithm, which was able to distinguish with a good performance among the most frequent IMIDs as indications for use. Considering that biological drugs are approved for multiple IMIDs, which may co-occur, and the variation in baseline risks among indications, accurate identification of their specific indications is essential to explore, for instance, their comparative safety. Thus, applying this META-algorithm to Italian claims data may facilitate post-marketing surveillance of biological drugs such as TNF-alpha inhibitors, anti-interleukin, anti-integrins, and selective immunosuppressants in specific therapeutic areas. Given the widespread use of ICD-9-CM and ATC terminologies in claims data, the proposed META-algorithm is expected to be easily adapted to longitudinal electronic healthcare databases from other countries.
Ethics Approval
This study was conducted in the context of the multiregional active pharmacovigilance VALORE project, funded by the Italian Medicines Agency. This project was approved by the Ethical Committee of the Academic Hospital of Verona, according to the current national law.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
G.T. participated in advisory boards and seminars as a lecturer on topics not related to the paper and sponsored by the following pharmaceutical companies in the last two years: Eli Lilly; Sanofi; Amgen; Novo Nordisk; Sobi; Gilead; Celgene; Daiichi Sankyo, Takeda and MSD. He is also the scientific coordinator of the pharmacoepidemiology team at the University of Verona and of the academic spin-off “INSPIRE srl” that carried out in the last two years observational studies/systematic reviews on topics not related to the content of this article and which were funded by PTC Pharmaceutics, Kyowa Kirin, Shionogi, Shire, Chiesi and Daiichi Sankyo. Y.I. is the CEO of the academic spin-off “INSPIRE srl”, which has received funding for conducting observational studies from contract research organizations (RTI Health Solutions, Pharmo Institute N.V.) and from pharmaceutical Companies (Chiesi Italia, Kyowa Kirin s.r.l., Daiichi Sankyo Italia S.p.A.). The authors report no other conflicts of interest in this work.
Acknowledgments
The authors have nothing to acknowledge.
Data Sharing Statement
All available data are included in the manuscript and in the Supplementary Materials.
Additional information
Funding
References
- Su CG, Lichtenstein GR. Influence of immunogenicity on the long-term efficacy of infliximab in Crohn’s disease. Gastroenterology. 2003;125(5):1544–1546. doi:10.1016/j.gastro.2003.05.009
- El-Gabalawy H, Guenther LC, Bernstein CN. Epidemiology of immune-mediated inflammatory diseases: incidence, prevalence, natural history, and comorbidities. J Rheumatol Suppl. 2010;85:2–10. doi:10.3899/jrheum.091461
- Williams JP, Meyers JA. Immune-mediated inflammatory disorders (I.M.I.D.s): the economic and clinical costs. Am J Manag Care. 2002;8(21 Suppl):S664–S681; quiz S682–S685.
- McInnes IB, Gravallese EM. Immune-mediated inflammatory disease therapeutics: past, present and future. Nat Rev Immunol. 2021;21(10):680–686. doi:10.1038/s41577-021-00603-1
- Trifirò G, Gini R, Barone-Adesi F, et al. The role of European healthcare databases for post-marketing drug effectiveness, safety and value evaluation: where does Italy stand? Drug Saf. 2019;42(3):347–363. doi:10.1007/s40264-018-0732-5
- Marcianò I, Ingrasciotta Y, Giorgianni F, et al. How did the introduction of biosimilar filgrastim influence the prescribing pattern of granulocyte colony-stimulating factors? Results from a multicentre, population-based study, from Five Italian Centres in the years 2009–2014. BioDrugs. 2016;30(4):295–306. doi:10.1007/s40259-016-0175-4
- Ingrasciotta Y, Giorgianni F, Bolcato J, et al. How much are biosimilars used in clinical practice? A Retrospective Italian Population-based study of erythropoiesis-stimulating agents in the years 2009–2013. BioDrugs. 2015;29(4):275–284. doi:10.1007/s40259-015-0132-7
- Trifirò G, Isgrò V, Ingrasciotta Y, et al. Large-Scale postmarketing surveillance of biological drugs for immune-mediated inflammatory diseases through an Italian distributed multi-database healthcare network: the VALORE project. BioDrugs. 2021;35(6):749–764. doi:10.1007/s40259-021-00498-3
- Canova C, Simonato L, Barbiellini Amidei C, et al. A systematic review of case-identification algorithms for 18 conditions based on Italian Healthcare Administrative Databases: a study protocol. Epidemiol Prev. 2019;43(4 Suppl 2):8–16. doi:10.19191/EP19.4.S2.P008.089
- Research C for DE and. Best practices for conducting and reporting pharmacoepidemiologic safety studies using electronic healthcare data sets. U.S. Food and Drug Administration; 2020. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/best-practices-conducting-and-reporting-pharmacoepidemiologic-safety-studies-using-electronic. Accessed May 27, 2022.
- Bartolini C, Roberto G, Girardi A, et al. Validity of Italian administrative healthcare data in describing the real-world utilization of infusive antineoplastic drugs: the study case of rituximab use in patients treated at the University Hospital of Siena for onco-haematological indications. Front Oncol. 2023;13:1059109. doi: 10.3389/fonc.2023.1059109
- Spini A, Rosellini P, Bellan C, et al. Development and validation of a case-finding algorithm for the identification of non-small cell lung cancers in a region-wide Italian pathology registry. PLoS One. 2022;17(6):e0269232. doi:10.1371/journal.pone.0269232
- Conway G, Velonias G, Andrews E, Garber JJ, Yajnik V, Ananthakrishnan AN. The impact of co-existing immune mediated disease on phenotype and outcomes in inflammatory bowel diseases. Aliment Pharmacol Ther. 2017;45(6):814–823. doi:10.1111/apt.13940
- Ingrasciotta Y, Jin Y, Foti SS, et al. Real-world patient characteristics and use of disease-modifying anti-rheumatic drugs in patients with rheumatoid arthritis: a cross-national study. Clin Rheumatol. 2023;42(4):1047–1059. doi:10.1007/s10067-022-06478-4
- Macca L, Li Pomi F, Ingrasciotta Y, Morrone P, Trifirò G, Guarneri C. Hidradenitis suppurativa and psoriasis: the odd couple. Front Med. 2023;10:1208817. doi:10.3389/fmed.2023.1208817
- Asgari MM, Wu JJ, Gelfand JM, et al. Validity of Diagnostic Codes and Prevalence of Psoriasis and Psoriatic Arthritis in a Managed Care Population, 1996–2009. Pharmacoepidemiol Drug Saf. 2013;22(8):842–849. doi:10.1002/pds.3447
- Dobson-Belaire W, Goodfield J, Borrelli R, Liu FF, Khan ZM. Identifying psoriasis and psoriatic arthritis patients in retrospective databases when diagnosis codes are not available: a validation study comparing medication/prescriber visit-based algorithms with diagnosis codes. Value Health. 2018;21(1):110–116. doi:10.1016/j.jval.2017.06.012
- Kim SY, Servi A, Polinski JM, et al. Validation of rheumatoid arthritis diagnoses in health care utilization data. Arthritis Res Ther. 2011;13(1):R32. doi:10.1186/ar3260
- Convertino I, Cazzato M, Giometto S, et al. Validation of algorithms for selecting rheumatoid arthritis patients in the Tuscan healthcare administrative databases. Sci Rep. 2021;11(1):20314. doi:10.1038/s41598-021-98321-0
- Ingrasciotta Y, Spini A, L’Abbate L, et al. (2024). Comparing clinical trial population representativeness to real-world users of 17 biologics approved for immune-mediated inflammatory diseases: An external validity analysis of 66,639 biologic users from the Italian VALORE project. Pharmacological Research. 2024;200:107074. doi:10.1016/j.phrs.2024.107074
- Salles G, Barrett M, Foà R, et al. Rituximab in B-cell hematologic malignancies: a review of 20 years of clinical experience. Adv Ther. 2017;34(10):2232–2273. doi:10.1007/s12325-017-0612-x
- Roberto G, Spini A, Bartolini C, et al. Real word evidence on rituximab utilization: combining administrative and hospital-pharmacy data. PLoS One. 2020;15(3):e0229973. doi:10.1371/journal.pone.0229973
- Ingrasciotta Y, Isgrò V, Foti SS, et al. Testing of coding algorithms for inflammatory bowel disease identification, as indication for use of biological drugs, using a claims database from Southern Italy. Clin Epidemiol. 2023;15:309–321. doi:10.2147/CLEP.S383738
- Pezzolo E, Ciampichini R, Cazzaniga S, Sampietro G, Zucchi A, Naldi L. Psoriasis severity matters when dealing with all-cause mortality in psoriasis patients: a record linkage analysis in Northern Italy. Arch Dermatol Res. 2021;313(4):255–261. doi:10.1007/s00403-020-02101-1
- Perrone V, Giacomini E, Sangiorgi D, et al. Treatment pattern analysis and health-care resource consumption on patients with psoriatic arthritis or ankylosing spondylitis treated with biological drugs in a Northern Italian Region. Ther Clin Risk Manag. 2020;16:509–521. doi:10.2147/TCRM.S248390
- Degli Esposti L, Perrone V, Sangiorgi D, et al. Therapeutic strategies utilization and resource consumption in patients treated for psoriatic arthritis: findings from a real-world analysis in an Italian setting. Patient Prefer Adherence. 2019;13:187–194. doi:10.2147/PPA.S178603
- Perrone V, Losi S, Rogai V, et al. Treatment patterns and pharmacoutilization in patients affected by rheumatoid arthritis in Italian Settings. Int J Environ Res Public Health. 2021;18(11):5679. doi:10.3390/ijerph18115679
- Kirby JS, Miller JJ, Adams DR, Leslie D. Health care utilization patterns and costs for patients with hidradenitis suppurativa. JAMA dermatol. 2014;150(9):937–944. doi:10.1001/jamadermatol.2014.691
- French DD, Margo CE. Postmarketing surveillance rates of uveitis and scleritis with bisphosphonates among a national veteran cohort. Retina. 2008;28(6):889–893. doi:10.1097/IAE.0b013e31816576ef
- Klein NP, Ray P, Carpenter D, et al. Rates of autoimmune diseases in Kaiser Permanente for use in vaccine adverse event safety studies. Vaccine. 2010;28(4):1062–1068. doi:10.1016/j.vaccine.2009.10.115
- Khalsa A, Liu G, Kirby JS. Increased utilization of emergency department and inpatient care by patients with hidradenitis suppurativa. J Am Acad Dermatol. 2015;73(4):609–614. doi:10.1016/j.jaad.2015.06.053
- Dubreuil M, Peloquin C, Zhang Y, Choi HK, Inman RD, Neogi T. Validity of ankylosing spondylitis diagnoses in The Health Improvement Network. Pharmacoepidemiol Drug Saf. 2016;25(4):399–404. doi:10.1002/pds.3952
- Wang Q, Zhang H, Dai SM. Differentiating psoriatic arthritis sine psoriasis from seronegative rheumatoid arthritis-Experiences from five patients. Int J Rheum Dis. 2022;25(9):1088–1092. doi:10.1111/1756-185X.14385
- Holroyd CR, Seth R, Bukhari M, et al. The British Society for Rheumatology biologic DMARD safety guidelines in inflammatory arthritis. Rheumatology. 2019;58(2):e3–e42. doi:10.1093/rheumatology/key208
- Baumgart DC, Misery L, Naeyaert S, Taylor PC. Biological therapies in immune-mediated inflammatory diseases: can biosimilars reduce access inequities? Front Pharmacol. 2019;10:279. doi:10.3389/fphar.2019.00279
