Abstract
Minichromosome maintenance (MCM) proteins play an essential role in DNA replication. They have been shown to be overexpressed in various types of cancer. However, the role of this family in renal cell carcinoma (RCC) is widely unknown. In this study, we have identified a number of RCC datasets in the Gene Expression Omnibus database and also investigated the correlation between the expression levels of MCM genes and clinicopathological parameters. We found that the expression levels of MCM genes are positively correlated with one another. Expression levels of MCM2, MCM5, MCM6, and MCM7, but not of MCM3 and MCM4, were higher in RCC compared to paired adjacent normal tissue. Only the expression level of MCM4, but not of other MCMs, was positively correlated with tumor grade. In addition, a high-level expression of MCM2 in either primary tumor or metastases of RCC predicted a shorter disease-free survival time, while a high-level expression of MCM4 or MCM6 in primary tumor was also associated with poorer disease-free survival. Interestingly, we also demonstrated that patients with their primary RCC overexpressing 2 or more MCM genes had a shorter disease-free survival time, while those with RCC metastases overexpressing 3 or more MCM genes had a shorter disease-free survival. Importantly, we also demonstrated that overexpression of MCM genes is an independent predictor for survival in RCC patients. Our results suggest that MCM2–7 genes may be an important prognostic marker for patients with RCC.
Introduction
DNA replication is an essential step for cell division which requires stringent control. Minichromosome maintenance (MCM) proteins bind to the replication origins for proper DNA replication.Citation1 All eukaryotic organisms, including human, possess 6 highly related MCM genes – MCM2–7.Citation2 These genes have been shown to play an important role in cancer development and have been linked to various cancer hallmarks,Citation3 especially for enhanced proliferation in cancer. Indeed, MCM genes were dysregulated in many types of cancer. Previously, we have shown that all MCM genes are overexpressed in breast cancer and that the number of MCM genes expressing at a high level is a more powerful prognostic marker in breast cancer than any individual MCM gene.Citation4
Renal cell carcinoma (RCC) accounts for around 90% of kidney cancer, and when diagnosed early, it can be successfully managed by surgery.Citation5 However, advanced unresectable and metastatic RCC is refractory to conventional chemotherapy, and vascular endothelial growth factor (VEGF) or mammalian target of rapamycin (mTOR) inhibitors become the major treatment options for this type of difficult-to-treat RCC.Citation5 Recently, immunotherapy has been an another option for the treatment of metastatic RCC with better survival and quality of life outcome demonstrated in patients treated with nivolumab compared to everolimus in a Phase III study.Citation6,Citation7 However, not all the metastatic RCC patients respond to these available therapies, and therefore, the identification of novel prognostic markers and potential therapeutic targets in RCC is still required.
MCM2 has been previously shown to be a proliferation marker,Citation8 and its expression is associated with high tumor grade and poor prognosis in RCC.Citation9 However, little is known about the expression of other MCM genes in RCC. In this study, we utilized the publicly available RCC patient datasets in Gene Expression Omnibus (GEO) database to investigate the expression of MCM2–7 genes in RCC.
Materials and methods
Extraction of clinical and microarray gene expression data from RCC patient datasets
RCC patient datasets were identified in the GEO database through keyword search for “Renal cell carcinoma”. This search was performed on March 18, 2017, with 908 returns. Datasets with >100 kidney specimens, available clinicopathological parameters, available probes, and expression data for MCM genes were included in this study; These datasets are GSE19949,Citation10 GSE40435,Citation11 GSE46699,Citation12,Citation13 and GSE53757.Citation14 GSE22541Citation15 was also included as it is the largest RCC patient dataset with disease-free survival status. Microarray gene expression data were retrieved from the data matrixes deposited to the GEO database by the original authors. Briefly, the GEO website has standardized URLs for its individual datasets, and for each of them, links are provided to the Series Matrix File(s) comprising the expression values for each of the probeset for each sample. Gene expression data were then retrieved from the matrices. R scripting was used to extract the expression values of a small number of genes (probesets) of interest, and the clinical data from the data matrixes were downloaded from GEO as described earlier.Citation16
Correlations of gene expression levels and clinical data
All statistical analyses were performed using SPSS19.0. Difference in expression levels between normal and cancer specimens was examined by Welch’s t-test. Correlation between the expression levels of different MCM genes and that between the expression levels of MCM genes and tumor grade were examined by Spearman’s rank test. The expression levels of MCM genes were divided into high and low levels using median expression level as the cutoff point for Kaplan–Meier survival analysis. Results were then compared by log-rank test. Multivariate analysis was performed by using Cox-regression model in which forward conditional stepwise approach was employed with a cut-off p-value of <0.05 as entry criteria. In all statistical tests, a p-value of <0.05 was considered as statistically significance.
Results
The correlation between the expression levels of MCM genes
As shown in to , expression levels of MCM2–7 genes were positively correlated with one another in the RCC specimens in all the 4 patient datasets analyzed (, GSE19949; , GSE40435; , GSE46699; , GSE53757). Most of these positive correlations were statistically significant. Especially for MCM2, its expression was consistently correlated with that of other MCM genes, including MCM4, MCM5, and MCM6, in the 4 datasets. These results suggest that expression of MCM genes may be tightly regulated within RCC.
Table 1 GSE19949 dataset of RCC specimens’ (n=113) MCM correlation
Table 2 GSE40435 dataset of RCC specimens’ (n=101) MCM correlation
Table 3 GSE46699 dataset of RCC specimens’ (n=63) MCM correlation
Table 4 GSE53757 dataset of RCC specimens’ (n=72) MCM correlation
The expression levels of MCM genes in normal kidney and RCC specimens
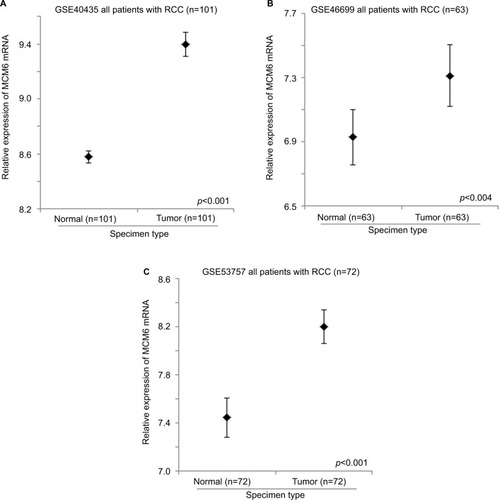
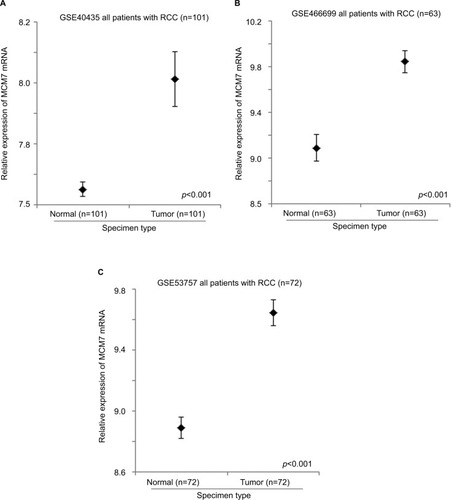
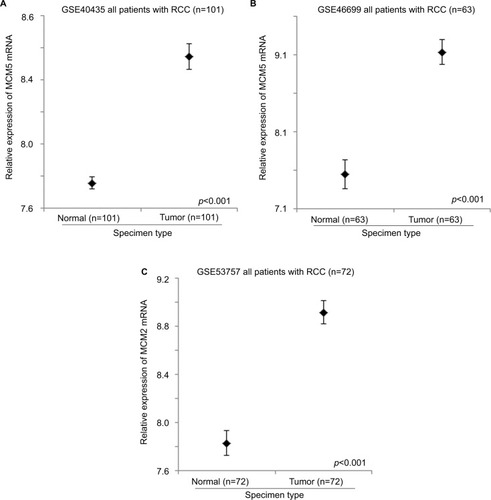
GSE40435, GSE46699, and GSE53757 datasets consist of 101, 63, and 72 pairs of normal kidney and RCC specimens. These datasets were used to compare the expression levels of MCM genes between normal kidney and RCC using paired t-test. The expression level of MCM2 gene was significantly higher in RCC specimens compared to normal kidney specimens in GSE40435 (; p<0.001), GSE46699 (; p<0.001), and GSE53757 (; p<0.001) patient cohorts. Similarly, the expression levels of MCM5 (), MCM6 (), and MCM7 () genes were significantly higher in RCC specimens compared to normal kidney specimens in all the 3 datasets. However, the expression level of MCM3 gene was higher in RCC compared to normal kidney specimens only in 2 datasets (GSE46699 and GSE53757) and was lower in RCC compared to normal kidney specimens in the other dataset (GSE40435). On the other hand, the expression level of MCM4 gene was significantly higher only in 2 datasets (GSE40435 and GSE53757), while the differential expression of MCM4 gene in GSE46699 was not statistically significant. Our results showed that not all the MCM genes were overexpressed in RCC compared to normal kidney and that MCM2, 5, 6, and 7 genes may play a more important role in the initiation of RCC.
Figure 1 The expression level of MCM2 mRNA in normal and RCC specimens. Error bar plots for the expression level of MCM2 mRNA in normal and RCC specimens in (A) GSE40435, (B) GSE46699, and (C) GSE53757 datasets.
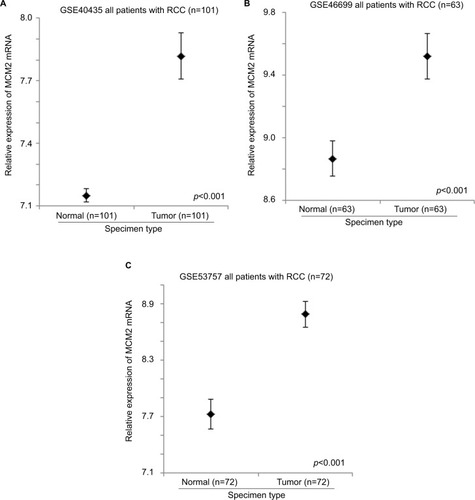
Figure 2 The expression level of MCM5 mRNA in normal and RCC specimens. Error bar plots for the expression level of MCM5 mRNA in normal and RCC specimens in (A) GSE40435, (B) GSE46699, and (C) GSE53757 datasets.
The expression levels of MCM genes in RCC specimens with different tumor grades
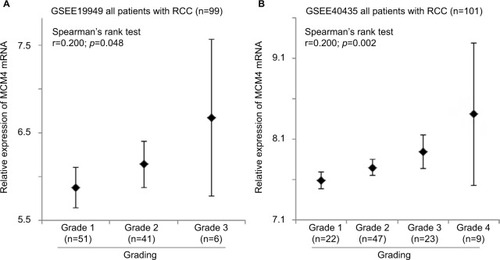
Tumor grade was available in both GSE19949 and GSE40435 datasets. The correlation between the expression levels of MCM2–7 genes and tumor grade was investigated in these 2 datasets. As shown in , only the expression level of MCM4 gene was significantly positively correlated with tumor grade in both GSE19949 (; r=0.200, p=0.048) and GSE40435 (; r=0.301, p=0.002). Although MCM4 was positively correlated with tumor grade, the correlations between tumor grade and the expression levels of MCM2 and MCM6 were significantly detected in 1 dataset but not in the other. Our results suggest that the expression of MCM4 gene may play a more prominent role in predicting tumor grade in RCC.
Figure 5 The expression level of MCM4 mRNA in RCC specimens with different grade. Error bar plots for the expression level of MCM4 mRNA in RCC specimens with different grade in (A) GSE19949 and (B) GSE40435 patient datasets. The Fuhrman system is used for grading renal cancer in both datasets analyzed.
The prognostic significance of MCM genes in RCC
Patient disease-free survival was available in GSE22541 with the expression levels of MCM2–7 gene also available in both primary and metastases specimens. Therefore, the association between the expression levels of MCM genes and disease-free survival was investigated in this dataset.
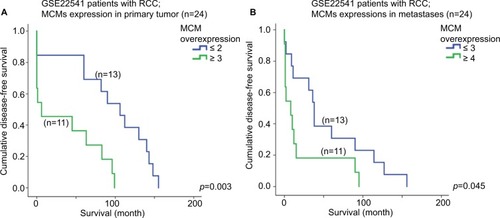
As shown in , a high expression level of MCM2 gene in the primary tumor of RCC was significantly associated with a shorter disease-free survival time (p=0.014); patients with their RCC primary tumor expressing a high level of MCM2 gene had a median disease-free survival time of 31.5 months (95% CI =1.9–61.1 months), while patients with RCC primary tumor expressing a low level of MCM2 gene had a median disease-free survival time of 85.4 months (95% CI =59.1–111.7 months).
Figure 6 Survival analysis of individual MCM gene. Kaplan–Meier survival curves for the expression of MCM2 gene in RCC (A) primary tumor and (B) metastases for disease-free survival in GSE22541 dataset. Kaplan–Meier survival curves for the expression of (C) MCM4 and (D) MCM7 gene in RCC primary tumor for disease-free survival in GSE22541 dataset.
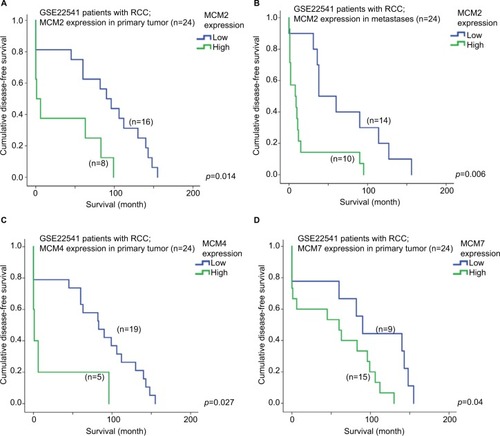
Interestingly, a high expression level of MCM2 gene in RCC metastases was also significantly associated with a shorter disease-free survival time (; p=0.006); patients with their RCC metastases expressing a high level of MCM2 gene had a median disease-free survival time of 18.4 months (95% CI =1.7–35.0 months), while patients whose RCC metastases were expressing a low level of MCM2 gene had a median disease-free survival time of 69.0 months (95% CI =37.9–111.7 months).
Other than MCM2 genes, we found that the expression levels of MCM4 and MCM7 genes in RCC primary tumor were both significantly associated with disease-free survival in this RCC patient cohort. As shown in , patients with a high level of MCM4 expression in their RCC primary tumor had a median disease-free survival time of 20.6 months (95% CI =0.0–57.0 months), which was significantly shorter compared to a median disease-free survival time of 79.8 months (95% CI =56.0–103.6 months) for those patients with a low level of MCM4 expression in their RCC primary tumor (p=0.027). Similarly, patients with a high level of MCM7 expression in their RCC primary tumor had a median disease-free survival time of 53.4 months (95% CI =28.7–78.1 months), which was significantly shorter compared to a median disease-free survival time of 90.9 months (95% CI =50.8–130.9 months) for those patients with a low level of MCM4 expression in their RCC primary tumor (; p=0.040).
The associations between the expression levels of other MCM genes, when considered individually, and disease-free survival were not statistically significant. However, when the MCM2–7 genes were considered together, a significant association between their expression levels, either in RCC primary tumor or in metastases, and disease-free survival was revealed. As shown in , patients with their RCC primary tumor expressing 2 or more MCM2–7 genes at a high level had a median disease-free survival time of 35.7 months (95% CI =10.7–60.7 months), which was significantly shorter than the median disease-free survival time of 94.3 months (95% CI =65.8–122.8 months) for patients whose RCC primary tumor were expressing less than 2 MCM2–7 genes at a high level (p=0.003). Similarly, patients with their RCC metastases expressing 3 or more MCM2–7 genes at a high level had a median disease-free survival time of 21.4 months (95% CI =0.35–42.4 months), which was significantly shorter than the median disease-free survival time of 54.8 months (95% CI =26.9–82.6 months) for patients whose RCC metastases were expressing <3 MCM2–7 genes at a high level (; p=0.045). By multivariate analysis, with additional parameters including T-stage, N-stage, and grade being input into the system, we found that both N-stage (p=0.003; 95% CI =2.770–144.998) and MCM expressions (p=0.002; 95% CI =1.933–20.447) were independent predictors of survival for RCC.
Discussion
In this study, we have found that the expression levels of MCM2–7 genes were positively correlated with one another in RCC specimens. MCM2, MCM5, MCM6, and MCM7 expressions were significantly upregulated in RCC tumor specimens compared to normal kidney specimens. Individually, MCM2, MCM4, or MCM7 expression was significantly associated with disease-free survival in primary RCC. When the expression levels of MCM2–7 genes were considered together, a significant association between the number of MCM2–7 genes expressing at high level, either in primary or metastases specimens, and disease-free survival were revealed. Our results suggest that the expression of MCM genes play an important role in predicting prognosis in RCC patients.
Little is known about the expression of MCM genes in RCC. MCM2 has been shown to be a proliferative and prognostic marker in RCC by 2 independent research groups.Citation9,Citation17 The dataset used in this study was able to provide similar results as earlier from others showing that MCM2 is a prognostic marker in RCC. In a cross search with MCM3–7 and RCC/kidney cancer in PubMed, only 2 RCC/MCM-related publication returned. One report showed that MCM3 was overexpressed in multiple cancers, including kidney cancer,Citation18 while the other one demonstrated that MCM7 may play a role in SETD2 loss-of-function mutation in RCC.Citation19 The limited search results indicate the importance of our findings as the expression of MCMs other than MCM2 may not be clearly described in the literature.
Indeed, several research reports have been investigating multiple MCM genes in a single study including breast,Citation16 glioma,Citation20 and pancreatic cancer.Citation21 To the best of our knowledge, this is the first report investigating the expression levels of MCM2–7 in RCC, showing that multiple MCM genes are overexpressed in RCC compared to paired normal specimens and the prognostic significance of MCM2–7 genes, when considered together, in RCC. As MCM2–7 genes have been shown to play a significant role in multiple types of cancer, with the current report showing that the expressions of MCM2–7 were significantly associated with RCC and could be used as a predictor for poor prognosis in this cancer type, the mechanism of how MCM2–7 genes promote RCC development and progression warrant further investigation in vitro and in vivo. In addition, whether expression of MCMs could predict treatment response should also be investigated. Since RCC patients with high-level expression of MCMs had a poorer survival, these patients may have limited response to available treatments as they have been treated in a curative intent. As a result, novel therapeutic strategies may be tested in this group of patients with a higher medical unmet need in upcoming clinical trials.
However, there are limitations in the current studies which include its retrospective nature, small sample size, and also the availability of clinicopathological parameters in online datasets varies, a prospective well-powered study may be required to demonstrate the potential clinical utility of MCMs as a prognostic marker in RCC.
In conclusion, our study shows that MCM genes may play an important role in RCC expression of MCM2–7 genes which may be considered together as a prognostic marker for RCC patients.
Acknowledgments
This study was supported by the Science and Technology Development Fund of Macau SAR (FDCT) (018/2015/A1) and the Health Science Research Personnel Training Program of Fujian Province (2017-CXB-22). The authors would like to thank Dr Ravikiran Mahadevappa for his technical assistance in this study.
Disclosure
The authors report no conflicts of interest in this work.
References
- KearseySELabibKMCM proteins: evolution, properties, and role in DNA replicationBiochim Biophys Acta199813981131369689912
- JenkinsonERChongJPMinichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insertProc Natl Acad Sci U S A20061037613761816679413
- NevesHKwokHFIn sickness and in health: The many roles of the mini-chromosome maintenance proteinsBiochim Biophys Acta2017186829530828579200
- KwokHFZhangSDMcCruddenCMPrognostic significance of minichromosome maintenance proteins in breast cancerAm J Cancer Res20155527125628920
- HsiehJJPurdueMPSignorettiSRenal cell carcinomaNat Rev Dis Primers201731700928276433
- MotzerRJEscudierBMcDermottDFNivolumab versus everolimus in advanced renal-cell carcinomaN Engl J Med20153731803181326406148
- CellaDGrunwaldVNathanPQuality of life in patients with advanced renal cell carcinoma given nivolumab versus everolimus in CheckMate 025: a randomised, open-label, phase 3 trialLancet Oncol201617994100327283863
- MehdiMZNagiAHNaseemNMCM - 2 and Ki - 67 as proliferation markers in renal cell carcinoma: a quantitative and semi – quantitative analysisInt Braz J Urol2016421121112827532114
- DudderidgeTJStoeberKLoddoMMcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinomaClin Cancer Res2005112510251715814627
- BeleutMZimmermannPBaudisMIntegrative genome-wide expression profiling identifies three distinct molecular subgroups of renal cell carcinoma with different patient outcomeBMC Cancer20121231022824167
- WozniakMBLe Calvez-KelmFAbedi-ArdekaniBIntegrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United StatesPLoS One20138e5788623526956
- Eckel-PassowJESerieDJBotBMSomatic expression of ENRAGE is associated with obesity status among patients with clear cell renal cell carcinomaCarcinogenesis20143582282724374825
- Eckel-PassowJESerieDJBotBMANKS1B is a smoking-related molecular alteration in clear cell renal cell carcinomaBMC Urol2014141424479813
- von RoemelingCARadiskyDCMarlowLANeuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4Cancer Res2014744796481024962026
- WuttigDZastrowSFusselSCD31, EDNRB and TSPAN7 are promising prognostic markers in clear-cell renal cell carcinoma revealed by genome-wide expression analyses of primary tumors and metastasesInt J Cancer2012131E693E70422213152
- MahadevappaRNevesHYuenSMThe prognostic significance of Cdc6 and Cdt1 in breast cancerSci Rep2017798528428557
- RodinsKChealeMColemanNFoxSBMinichromosome maintenance protein 2 expression in normal kidney and renal cell carcinomas: relationship to tumor dormancy and potential clinical utilityClin Cancer Res200281075108111948116
- HaSAShinSMNamkoongHCancer-associated expression of minichromosome maintenance 3 gene in several human cancers and its involvement in tumorigenesisClin Cancer Res2004108386839515623617
- KanuNGronroosEMartinezPSETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repairOncogene2015345699570825728682
- HuaCZhaoGLiYBieLMinichromosome Maintenance (MCM) family as potential diagnostic and prognostic tumor markers for human gliomasBMC Cancer20141452625046975
- PengYPZhuYYinLDThe expression and prognostic roles of MCMs in pancreatic cancerPLoS One201611e016415027695057