Abstract
With the clinical promotion of precision medicine and individualized medical care, molecular targeted medicine has been used to treat non-small cell lung cancer (NSCLC) patients and proved to be significantly effective. Anaplastic lymphoma kinase (ALK) inhibitor is one of the most important specific therapeutic agents for patients with ALK-positive NSCLC. It can extend the survival of patients. However, resistance to the ALK inhibitor inevitably develops in the application process. So, the real-time resistance surveillance is particularly important, and liquid biopsy is one of the most potential inspection methods. Circulating tumor cells, circulating free tumor DNA and exosome in body fluid are used as the main detection biomarkers to reflect the occurrence of resistance in real time through sequencing or counting and then to guide the follow-up treatment.
Video abstract
Point your SmartPhone at the code above. If you have a QR code reader the video abstract will appear. Or use:
Introduction
As a malignant disease, lung cancer ranks first in incidence and mortality of cancer, and has so for a long period of time. It is predicted that there still will be more than 220,000 new cases and more than 160,000 deaths in the US in 2017.Citation1 Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for about 80%–90%.Citation2–Citation4 However, more than 50% of newly diagnosed patients have been in advanced stage or have gained metastases, and thus they have lost the opportunity for surgical treatment. Radiotherapy or chemotherapy is not ideal for them.Citation5 Radiotherapy and chemotherapy are typical traditional tumor treatments but they lack cell killing specificity. This results in these treatments to not only kill tumor cells but also damage normal cells.. Therefore, they will produce a variety of toxic side effects on all systems in the body, and some patients cannot tolerate treatment. Though chemotherapy is still the main treatment for advanced NSCLC patients, clinical data suggest that the development of chemotherapy has encountered the situation of bottleneck.Citation6 It cannot further extend remission and survival. Radiotherapy could only serve as a palliative reduction therapy.Citation7 Furthermore, though radiotherapy and specific chemotherapy could be received by outpatients, compared with oral medication, they are considerably inconvenient, since patients who receive the treatment need to visit the hospital frequently. Due to the shortcomings of radiotherapy and chemotherapy, targeted therapy has gradually been paid more attention to and has gradually become a first-line treatment program.
With genetic and precision medicine developing in recent years, it has been found that the development of lung cancer is closely related to tumor-driven gene. Anaplastic lymphoma kinase (ALK) fusion gene and epidermal growth factor receptor (EGFR) are the NSCLC-driven genes that are currently being recognized.Citation8–Citation12 ALK fusion gene occurs in 3%–7% of patients with NSCLC, which includes echinoderm microtubule-associated protein-like 4-ALK (EML4-ALK), TRK-fused gene-ALK, nucleophosmin-ALK and kinesin family member 5B-ALK.Citation13–Citation18 ALK inhibitor (ALKi) is an effective targeted medicine for patients with ALK mutations, which can significantly prolong their survival. Crizotinib is a first-generation ALKi, and it has become a recommendation for the treatment of ALK-positive NSCLC patients in the guidelines of Europe and the US, and has been used as a first-line therapy for patients in advanced stage.Citation19,Citation20 Crizotinib can not only significantly prolong the progression-free survival (PFS), but also increase the objective response rate (ORR). The clinical trial of crizotinib (PROFILE 1014) indicated that the median PFS was 10.9 months, while the ORR was 74% and 1-year survival rate was 84%, demonstrating better efficacy than chemotherapy.Citation21 But before long, it was found that about 30% of patients who used crizotinib as initial treatment have primary resistance to it, some patients developed secondary resistance within 1–2 years and about 40% developed metastasis in the central nervous system.Citation22
With the emergence of crizotinib resistance, the second- and third-generation ALKi were gradually introduced and approved for clinical application. It was observed that the ORR and the median PFS of crizotinib-resistant patients increased to 50% and 8.9 months, respectively, after being treated with alectinib, a second-generation ALKi.Citation23 The third-generation ALKi lorlatinib had a strong inhibitory effect on known resistant mutations and had a good therapeutic effect on ALK-driven brain metastasis.Citation24 The ongoing I/II trial shows that the ORR of lorlatinib is 26%, indicating that the timely changing of treatment program can still bring about a positive effect when the original treatment escapes.Citation25 But the direct application of the latest and most effective ALKi is not equal to permanent efficacy and benefit. A study has showed that a patient with lorlatinib escape had a new tumor gene mutation that caused his re-sensitization to crizotinib.Citation26 All of these show that it is of clinical significance to guide the treatment at the molecular level after resistance. The real-time detection technology that can be applied to cancer patients is needed urgently in clinical practice. At present, the regular surveillance of lung cancer during treatment mainly depends on the pathological diagnosis after puncture biopsy and imaging examination. But pathological biopsy is an invasive operation that cannot be carried out in all patients, and the compliance of the patients becomes worse during repeated operation. Furthermore, the error resulting from tumor heterogeneity cannot be overcome now, while the imaging examination is often delayed.
As early as the 19th century, Paget had put forward the seed-soil theory.Citation27 It was thought that “seeds” would fall off from the primary tumor with the appropriate distal organs as “soil” to form metastases, which was the most original definition of what is called the circulating tumor cell (CTC) now. Subsequently, the researchers also found other tumor-derived biomarkers, such as circulating tumor DNA (ctDNA), exosome, etc. These biomarkers carry information about the source of the organization and flow freely in various body fluids.Citation28 Liquid biopsy distinguishes itself as a new technique for separation and analysis of the tumor-derived biomarkers in body fluid, which obtain the coding information of tumor. It is an ideal technique for ALK-positive NSCLC resistance surveillance for its advantages such as simplicity, minimally invasive, real time and repeatability.
The mechanisms of ALKI resistance
Despite the constant upgrading of ALKi and its enhancing efficacy, the resistance still cannot be overcome. ALKi resistance can be divided into primary resistance and secondary resistance.
Primary resistance
Primary resistance refers to the ineffective medical treatment in the early stage.Citation29 About 25% of patients treated with crizotinib have primary resistance.Citation30 Although the mechanism of primary resistance is not fully understood yet, it is speculated that it is closely related to ALK variations. ALK variations include the fusion of ALK with other partner genes encoding protein other than EML4 and structural variations of ALK itself.Citation31 Experiments showed that there is a significant difference in sensitivity to crizotinib among tyrosine kinase receptors coded by different ALK fusion genes or different subtypes of same ALK fusion gene.Citation32 In addition, tumor heterogeneity can also lead to primary resistance. Studies have found that 5% –8% of cancer cells in ALK-positive NSCLC patients contain EGFR mutation, which causes the failure of ALKi. Another cause may be false-positive genotype. ALK tumor-driven mutation can be monitored by a variety of techniques, and ALK fluorescence in situ hybridization (FISH) testing is considered the current gold standard procedure.Citation33–Citation35 It is believed that false-positive results could be led by artificial cutting gene fragment error, cell morphological differences, probe abnormalities or pathological misunderstanding in the detection process.Citation36 Even VENTANA ALK (D5F3) CDx assay with higher specificity and sensitivity in detecting ALK-positive NSCLC, which is recently approved by the US Food and Drug Administration (FDA), can also cause false-positive results.Citation37 Thus, in a few cases, ALKi primary resistance may be caused by technical factors rather than a real existence of biological resistance. Finally, although ALK FISH could recognize true ALK translocation, these patients did not have functional rearrangements, which is also a pseudo-positive resistance that results from pseudo-primary genotype.
Secondary resistance
Secondary resistance, also known as acquired resistance, refers to the recurrence of tumor progression that the original ALKi-sensitive patients suffer after complete remission or partial remission after more than 6 months with medical treatment.Citation38 Secondary resistance can be divided into dominant and non-dominant. Dominant secondary resistance mainly refers to ALK kinase domain mutation (29%) and increase of ALK gene copy number (9%), which accounts for one-third of crizotinib resistance by increasing the activity of tyrosine kinases.Citation39–Citation41 Other types of resistance are non-dominant, including ALK signal bypass activation, tumor heterogeneity, etc. In fact, two or more resistance patterns could be generated in the same patient. A large part of the resistance mechanism still remains unknown.
Mutation in the target ALK gene
When the first mutation emerges in the ALK fusion gene, it induces the production of tumor; when secondary mutation comes up, it means the emergence of resistance. Most of the mutations currently found in the target ALK gene are mainly point mutation. C1156Y and L1196M were the first discovered mutant types, followed by L1152R, G1202R, G1269A, F1174L, 1151Tins, S1206Y, I1171T, D1203N, V1180L, etc.Citation32,Citation42,Citation43 Crizotinib is an ATP-competitive selective inhibitor of the ALK and MET tyrosine kinases, which inhibits their tyrosine phosphorylation.Citation44 With low level of tyrosine kinase phosphorylation, aberrant downstream pathways of ALK mutations will be inhibited. In general, these aberrant downstream pathways can lead to apoptosis insufficiency, excessive cell proliferation and the development of NSCLC.Citation45 Ultimately, crizotinib reaches the pharmacological effect of restraining the development of tumor. As the secondary mutation emerges, the protein composition of tyrosine kinase changes and its activity increases, which leads to the decrease of the inhibitory effect. But the point mutations do not destroy the effect of ALK signaling pathway, and so the tumor can still rely on the pathway for survival and development. Now the most common mutation of ALK kinase domain in the crizotinib resistance is L1196M, leading to replacement of leucine by methionine in the kinase region. It helps in the formation of the protein active conformation and increases the activity of protein kinase, interfering with the inhibitory effect of the targeted treatment on kinase activity, and finally leading to treatment failure.Citation46 Studies have shown that each ALKi will specifically correspond to a group of resistant mutant types. G1202R is one of the specific resistance mutation types that responses to the second generation of ALKi.Citation47
Abnormal copy number of fusion gene
ALK fusion gene copy number abnormalities include copy number gain (CNG) and gene deletion. The resistance mechanism of abnormal ALK gene copy number is similar to the gene mutation, increasing the activity of kinase. The CNG means that the average number of the rearranged genes in lung cells are more than triple. Katayama et al analyzed the genes in crizotinib-resistant patients and found that ALK fusion gene copy number increased remarkably, and confirmed that CNG is to blame for the resistance in vitro experiments.Citation41 Doebele et al happened to discover a special sample of resistance that may be caused by deletion of the ALK fusion gene.Citation46 It was after exclusion of common causes of resistance such as gene mutations, CNG, signal bypass activation and after multiple biopsies, that researchers thought that the resistance was indeed caused by genetic deficiency. But the view that ALK fusion gene deficiency is the mechanism of ALKi resistance has not been fully verified.
Emergence of bypass tracks
Signal bypass activation refers to the activation of other carcinogenic drivers. When ALKi inhibits the level of tyrosine kinase phosphorylation and blocks the downstream signaling pathways, tumor cells can reduce its dependence on ALK and downstream signals by activating other signaling pathways or the ALK signaling pathway in downstream that is not inhibited, resulting in the failure of ALKi. It results in 20% of the cases of crizotinib resistance.Citation49 The key signaling transduction pathways in downstream of ALK include Ras/MEK/ERK, PI3K/AKT and JAK3-STAT3.Citation50,Citation51 Takezawa et al found that ALKi could inhibit the amplification of EML4-ALK positive tumor cells and induce apoptosis by inhibiting the transduction pathways of ERK and STAT3, while the pathway of PI3K/AKT was not affected or less affected and finally causes resistance.Citation52 In addition, tumors can continue to develop by activating other tumor driven genes. The most common driven genes include EGFR and KRAS mutations, EGFR phosphorylation and c-KIT amplification.Citation39,Citation48 Activation of a variety of different carcinogenic drivers can occur simultaneously in the tumor. In an in vitro experiment, the expression of EGFR mRNA in cells with resistance was upregulated after applying crizotinib to ALK-positive cell lines, indicating that the continual progression of tumor could be owing to the activation of EGFR and persistent activation of downstream signal transduction pathway independent from ALK.Citation53
Tumor heterogeneity
There still lacks a thorough understanding about resistance on account of tumor heterogeneity. ALK-positive solid tumor could be a mixture of a majority of ALK-positive tumor cells and other tumor cells induced by different tumor-driven genes. The growth and amplification of predominant ALK-positive tumor cells would be inhibited and the original minority of tumor cells driven by other tumor genes was transformed into a dominant cell line after the application of ALKi. It is also possible that there are multiple dominant and non-dominant mechanisms of resistance in the tumor, for which ALKi would be helpless. The American Society of Clinical Oncology meeting reported a study on the distribution of driven genes in NSCLC patients in 2014. The study included 35 cases of patients with multiple pulmonary nodules, and it showed that the inconsistency rate of mutation-driven gene of different nodules reached 68.6%.Citation54 But the current mainstream surveillance techniques, such as pathological biopsy and imaging examination, still cannot overcome the detection error of tumor heterogeneity.
Others
In addition, there are some rare types of non-dominant resistance. Researchers had found an abdominal nodule transformed from small cell lung cancer (SCLC) in a patient with crizotinib treatment failure.Citation55 In another case, SCLC-transformed issue was detected in the lung tissue of alectinib-resistant patients.Citation56 They both indicated the potential of SCLC transformation to be a novel resistance mechanism. Moreover, ALKi resistance is probable to be achieved by epithelial-mesenchymal transition, autophagy, etc.Citation57,Citation58 The current discovered mechanisms are only a small part; many other mechanisms of resistance still need further study.
Liquid biopsy
Due to the insurmountable shortcomings of imaging and pathological biopsy methods, and the necessity for real-time resistance surveillance in the treatment of ALK-positive NSCLC patients, the liquid biopsy technology that can achieve minimally invasive and real-time surveillance has attracted more and more attention and expectation. The most common biomarkers of liquid biopsy are CTCs, ctDNA and exosome. They are permissible for separation, purification and analysis of tumor DNA, mRNA, miRNA and other genetic materials. Since biomarkers can be derived from different parts of the tumor or different cancer foci, liquid biopsy can overcome the interference of tumor heterogeneity and reflect the overall information of tumor. Therefore, liquid biopsy is the most potential and ideal detection method for resistance surveillance at present. The research progress of liquid biopsy in resistance surveillance is shown in .
Figure 1 The research progress of liquid biopsy in resistance surveillance.
Abbreviations: CTC, circulating tumor cell; ctDNA, circulating tumor DNA; NGS, next-generation sequencing; ddPCR, digital droplet polymerase chain reaction; CAPP-Seq, cancer personalized profiling by deep sequencing; EFIRM, electric field-induced release and measurement technology.
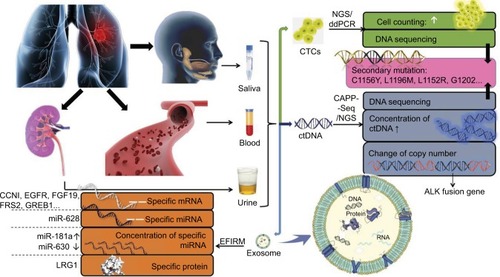
CTC
CTC refers to the tumor cells that fall off from primary lesions or metastases of tumor and then enter into the blood circulation.Citation59 But the detection rate of CTC is low; only one CTC can be detected in an average of 105–107 monocytes in the peripheral blood of patients with advanced tumors.Citation60 Genetic materials, such as DNA, RNA and protein, are contained in CTCs. Digital droplet polymerase chain reaction combined with next-generation sequencing (NGS) is currently used to detect and sequence genes in CTCs, which help to obtain the information about the source of the organization.Citation61 CTC can also be detected by CellSearch, which is approved by FDA.Citation62 When secondary resistance occurs or other carcinogenic drivers are activated, theoretically, resistance can be indicated by the percentage of mutated CTCs and the mutations of gene, mRNA and protein. The half-life of CTC in blood is only a few hours. Therefore, it can be used as a real-time detection target.Citation63,Citation64
The mutation of L1196M was found by enrichment and gene sequencing of the CTCs in peripheral blood of patients with crizotinib secondary resistance, indicating that the resistance mutation can be found at an early stage through the gene sequencing on mutated CTCs.Citation65 Thus, it would be beneficial to conduct a large sample of gene sequencing to obtain the specific resistant mutation spectrum of different ALKi, which may be helpful for the diagnosis of resistance. In another study, researchers tried to obtain the quantitative change of EML4-ALK+ CTC detected in serum of patients with adenocarcinoma as a prompt of resistance.Citation66 At first, three CTCs were detected before the treatment with crizotinib, one of which was EML4-ALK + CTC. After treatment, no EML4-ALK+ CTC was found, while two EML4-ALK + CTC recurred in the further progress of tumor. The reappearance of EML4-ALK + CTC indicated that the patients acquired crizotinib resistance. As there was no gene detection on CTC for surveillance, researchers give priority to the dominant resistance when the reasons for resistant are unknown. So, ceritinib was used for treatment, but EML4-ALK + CTC and tumor progression were still detectable subsequently. At last, it was considered as non-dominant resistance, and had chemotherapy as adjuvant therapy. Although the method of CTCs counting for resistance surveillance is less accurate than gene sequencing, it is simpler. Coupled with the clinical and imaging performance, the resistance and its mechanism can be roughly determined. TKI of other target genes is often used in combination for treatment when the non-dominant resistance occurs, especially when combined with other tumor-driven genes or in the presence of signal bypass activation. A study based on Asians found that 18.6% of ALK-positive patients had EGFR mutations, while 3.9% of EGFR mutation patients had ALK fusion.Citation67 The combination of ALKi and EGFRi can achieve a better inhibitory effect than the use of each of the specific inhibitors alone. In addition, using the treatment-resistant ALKi, or a combination of heat shock protein inhibitors (HSP90) or chemotherapy with the treatment-resistant ALKi has a positive significance for patients.Citation21,Citation68
Despite the low concentration of CTCs in peripheral blood and a lack of detection technology with high sensitivity and specificity, CTC is ideal for detection and analysis, for its capability of overcoming the disturbance of tumor heterogeneity because it is derived from different parts of the solid tumor or different cancer foci. Moreover, CTC has a full set of tumor genetic information that can be used to study tumor phenotype and cellular function. Due to the presence of resistance, a series of biopsies is usually required to detect the progression of the tumor after the use of ALKi. Though it is possible to perform multiple biopsy tests on the patients technically, it will be followed by operational risk and poor tolerability of the frail patients. Now DNA in CTCs is considered to be a more ideal detection target for re-biopsy.Citation69–Citation71
ctDNA
Circulating free DNA (cfDNA) is defined as the tissue-specific DNA fragment that is released into the blood.Citation72 CtDNA refers to the cfDNA that is secreted by tumor cells or released after apoptosis and necrosis.Citation73 The level of ctDNA in tumor patients is positively correlated with tumor progression, and the half-life is generally about 2 hours, which can better reflect tumor burden and the effectiveness of the current treatment in real time than imaging examination.Citation74,Citation75 Secondly, ctDNA sequencing can be performed by the technique of NGS and cancer personalized profiling by deep sequencing (CAPP-Seq), with which the tumor resistance or the change of gene after progression can be found.Citation76,Citation77 It is also possible for it to reflect the dominant tumor cell population at different points in time, regardless of the heterogeneity.Citation78
In a study, a tracing blood test was performed on two advanced NSCLC patients with crizotinib treatment, and found that the ctDNA in serum and the allele frequency of ALK rearrangements in ctDNA significantly increased after 10 months of medication, suggesting that the treatment failed.Citation79 Three resistance mutations, L1152R, I1171T and L1196M, could be detected in the ctDNA of one of the patients, indicating that the change of nucleic acid sequence and blood concentration of ctDNA can be sensitive to detect the resistance. At this point, the second generation of ALKi should be put into use. The experimental data showed that the ORR of the second-generation ALKi, ceritinib and alectinib, after the treatment with crizotinib failed, were 56% and 50% respectively.Citation22,Citation80 Especially, alectinib had a significant effect on secondary resistance caused by L1196R.Citation36 The other patient with central nervous system metastasis who had not used crizotinib in the same study sequentially used the second-generation ALKi ceritinib and alectinib, and had progress of metastases 1 year later.Citation81 The ALK G1202R mutation was detected in ctDNAs. The clinical manifestations and the results of ctDNA detection pointed out the occurrence of resistance, and then the patient switched to the third-generation ALKi lorlatinib. The clinical symptoms caused by metastases improved, and the concentration of ctDNA of G1202R in peripheral blood also decreased, showing the significance of ctDNA in resistance surveillance and guiding follow-up therapy. It is particularly important to detect ctDNA mutation after using the second-generation ALKi resistance. The second-generation ALKi can be used for patients who had or did not have the resistant mutation of the first-generation ALKi. While the presence of new secondary resistance mutation in ALK fusion gene, such as G1202R, is an essential indication of lorlatinib, the third-generation ALKi, otherwise, lorlatinib will not work.Citation82
Because ctDNA is freely available in the body fluids with low concentration, it is easily decomposed by enzymes or phagocytosed by cells. There still lacks effective separation and analysis technique at present. The current detection technique is prone to false-negative or false-positive results. Moreover, ctDNA is dissociated from the extracellular region, resulting in the detection result of nucleic acid not being co-located with protein, nor can the function of cells be studied. CtDNA is now the most commonly used biomarker in all reports of ALK-positive NSCLC resistance surveillance. But the testing standard and evaluation criteria are different in different laboratories. Therefore, it is necessary to provide a unified industry measurement standard before putting it into clinical use. But overall, ctDNA is indeed an ideal biomarker. It has the characteristics of minimally invasive, strong specificity and can overcome the tumor heterogeneity and reflect the integral information of the tumor. It is more sensitive to the detection of the mutation of the tumor and permissible for recognizing known resistance mutations. All of these traits meet the requirements of a repeatable biopsy technique that is urgently needed in the resistance surveillance process.Citation83–Citation85
Exosome
Exosome is an extracellular vesicle with a diameter of 30–120 nm, the surface of which is made up of lipid bilayer vesicles, containing DNA, mRNA, miRNA, protein and other genetic materials.Citation86,Citation87 In physiological and pathological conditions, exosome can be secreted by different types of cells.Citation88 It carries different components closely based on its parental-derived cells. The specific components can be referred in the “exosome content database” (www.exocarta.org).Citation89 The current exosome content database contains approximately 1,639 mRNAs, 764 miRNAs, 4,563 proteins and 194 lipids. The peripheral blood of tumor patients has higher concentration of exosome than that in normal ones, and it carries tumor-specific molecular substances such as DNA and gene mutations.Citation90 Although changes in protein-coding genes are key targets for elucidating tumor-specific changes, recent studies have shown that miRNAs have a greater advantage in detecting convenience and demonstrating tumor specificity. A study about ovarian cancer found that 8 miRNA-specific diagnostic markers could be detected simultaneously in tumor tissue and exosome of patients.Citation91 The miRNAs of exosome were introduced into the exosomes through a special sorting mechanism, implying that the exosome miRNA spectrum was highly similar to the miRNA spectrum of the parental cancer cells.Citation92–Citation94 Its information is more tumor-specific and better than global mRNA spectrum, which is used to define the types of cancer and other subtypes of RNA.Citation95,Citation96 Although there is no report illustrating the use of exosome in resistance surveillance currently, similar to tumor DNA, changes in copy number of exosome miRNAs and nucleic acid sequences are considered to be available for resistance surveillance.
Though the specific research of exosome miRNAs in the field of resistance surveillance is deficient temporarily, we can find its potential and future development according to the appliance of tumor miRNA. MiRNA is a small non-coding RNA, consisting of about 18–25 nucleotides and being responsible for negative regulatory gene expression after transcription. Specific miRNA is able to indicate its binding effect on proto-oncogenes or tumor suppressor genes.Citation97,Citation98 Studies have found that the expression of tumor miRNAs is associated with tumor biology. When the expression of certain miRNAs increases, it may lead to downregulation of tumor suppressor genes, while the decrease in expression of other miRNAs can lead to an increase in the expression of oncogenes.Citation99,Citation100 Both of them can lead to tumor progression and resistance. The sensitivity of NSCLC to cisplatin is associated with the upregulation of miR-181a, while the upregulation of miR-630 is related to resistance.Citation101 However, there is still a lack of research on the correlation between miRNA expression and ALKi resistance.
The abnormal expression of miRNA is a common feature of dysplasia and cancer. The expression profile of miRNA is associated with the progression of disease and prognosis.Citation102,Citation103 A total of 12 special miRNAs were found to be particularly present in the exosomes of patients with NSCLC, indicating that the current technique is capable of supporting miRNAs’ detection that were produced after resistance.Citation104 MiR-628, another marker in crizotinib-resistant cell line, shows an overexpression, indicating the significance of miRNA in resistance surveillance.Citation105
As with CTCs and ctDNA, exosomes all have important implications in resistance testing, and it may be more valuable than the former two. The concentration of exosomes in peripheral blood is higher than that of CTCs, and it can be detected in urine, saliva, cerebrospinal fluid, semen, milk, pleural effusion, ascites and other body fluids in addition to serum or plasma.Citation106 The exosome contents are protected by the vesicle membrane. They can be prevented from being destroyed by enzymes in the body fluids and become more complete and stable. The surveillance of platelet endocytosis of tumor-derived RNA is even earlier than imaging to detect tumor progression.Citation107
Other body liquid
Currently, liquid biopsy used in clinical and laboratory applications primarily uses peripheral blood to enrich and analyze the biomarkers. In fact, studies have proved that urine, saliva and pleural effusion are available for noninvasive detection.
Urine
A total of 150 patients with positive EGFR gene mutation (that is, L858R or L861Q positive) and EGFR-tyrosine kinase inhibitor (TKI) treatment accepted a long-term surveillance.Citation108 The data showed that the level of cfDNA in plasma and urine was significantly decreased in early treatment. With the application of TKI, resistant individuals gradually appeared. In all, 53% of the treatment-resistant patients had double growth of the cfDNA in plasma and urine. The secondary mutation EGFR T790M can be detected in some patients. Both liquid biopsy and pathologic biopsy were conducted to detect patients with resistance in this study. When urine was used as the detection sample of liquid biopsy, the detection results of liquid biopsy and pathologic biopsy overlapped by 88%, while the detection results of both biopsy techniques were exactly the same when using plasma samples for liquid biopsy. It indicated that urine could also serve as a liquid biopsy material, and urine cfDNA could be used as an indicator of tumor progression. In addition, comparing exosome protein in urine and lung cancer tissue of the normal control group with that of NSCLC patients, it was found that leucine-rich α-2-glycoprotein (LRG1) was highly expressed in urinary exosome and lung tissues of NSCLC patients, showing that the exosome LRG1 in urine may be a candidate biomarker for noninvasive diagnosis of NSCLC.Citation109
Saliva
With the development of real-time quantitative PCR detection system, the five biomarkers (CCNI, EGFR, FGF19, FRS2 and GREB1) of salivary exosome mRNA are effective in distinguishing lung cancer patients from normal controls.Citation110 These salivary mRNA biomarkers have the ability to detect lung cancer. Electric field-induced release and measurement technology (EFIRM) is also a popular technique for the analysis of exosomes currently. It can effectively cleave exosomes; release proteins, RNA and DNA; and detect tumor biomarkers.Citation111 At present, this technique has been proved to succeed in detecting tumor-driven mutations in saliva of NSCLC patients.Citation112 The difference between saliva detection and plasma detection is not significant.Citation113 It is expected to be used in diagnosis and treatment guidance.
Pleural effusion
EGFR mutation has been detected in the CTCs derived from pleural effusion, indicating that biomarkers from pleural effusion can offer a clue to the existence or progression of tumor.Citation114 Pleural effusion may be more specific than plasma, urine and saliva for its proximity to cancer foci. However, pleural effusion sampling, like tissue biopsy, is an invasive operation, which restricts its development.
The value of other body fluids on the diagnosis of NSCLC has been fully supported according to the current experimental results, of which there are a great many studies on EGFR-TKI resistance surveillance. More studies and experiments are expected to confirm their viability in ALKi resistance surveillance.
Discussion and conclusion
This paper focused on the use of liquid biopsy on ALKi resistance surveillance. Actually, liquid biopsy is applicable to the surveillance of all lung cancer patients. Although targeted therapy is superior to chemotherapy in PFS and tolerance, targeted therapy has not surpassed chemotherapy in terms of overall survival.Citation115 Patients with genetic mutations are a minority after all, and chemotherapy and radiotherapy are still the treatment of most patients with lung cancer.Citation116 Liquid biopsy can predict tumor progression, which can also guide the adjustment of chemotherapy program or increase the radiotherapy dose. Certainly, the best indication for liquid biopsy is targeted therapy, which is mainly for NSCLC patients. The existing clinical data point out that the molecular targeted medicine has a significant effect on NSCLC patients with gene mutation. Although it is not possible to solve the problem of resistance up to now, liquid biopsy plays an important role in warning resistance as well as its related mutation in early stage and guiding the alteration of treatment program, which has a positive effect on relieving the disease and prolonging the survival. With the further study of NSCLC, more and more specific tumor-driven genes and the corresponding targeted medicine are bound to be found. Liquid biopsy will keep promoting the development of targeted therapies.
Liquid biopsy is superior to traditional pathologic biopsy and imaging for its unique noninvasive nature, convenience and repeatability. However, many obstacles need to be overcome before it becomes a practical method to monitor treatment resistance. With small size and low concentration, there is still a lack of specific and sensitive separation and analysis techniques for the biomarkers used in the experimental study. The results of these studies from different laboratories also lack unified interpretation for resistance evaluation. It is prone to be false negative or false positive when using FISH, in situ hybridization (ICH) and other detection technology in disease surveillance, and their ratio of signal-to-noise tends to be low when detecting the early disease. The application of exosomes in resistance surveillance is still to be developed.
To overcome the obstacles of using liquid biopsy in clinical research and clinical practice in ALK-translocated patients, researchers came up with several solutions. Setting ALK-CNG as an example, an enrichment technique is supposed to be performed before detecting ALK-CNG, such as isolation by size of epithelial tumor cells, which is more effectively in CTC recovery than CellSearch.Citation117 Then, Pailler et al developed filter-adapted FISH to increase cell recovery as well as to detect mutations in ALK-positive CTCs.Citation118 For ctDNA, FISH is generally available in pathology laboratories for gene sequencing, but it is often used to identify copy number alternation of known gene.Citation119 A capture-based NGS ctDNA detection method called CAPP-Seq was developed with ultrasensitivity for even unknown genes and all major classes of mutations in large portions of the genome, while NGS were developed with ultrasensitive for specific genomic positions.Citation120 EFIRM is a newly developed technique that combines the rapid extracellular vesicle lysis procedure and detection and capture of molecular content from the EV with high sensitivity, which reduces the content degradation caused by exposure.Citation121 Though it is impossible to totally overcome false-negative and false-positive results in theory, it is advised to apply two kinds of detection techniques simultaneously to improve accuracy. For example, a combination of ICH assay and FISH is usually used in clinical research. ICH is usually recommended as a primary assay because of its greater sensitivity and specificity, while FISH is reserved for use only in ICH equivocal cases.Citation119,Citation122 To date, the absence of unified interpretation for resistance evaluation results from the standpoints varying from one researcher to another and different detection techniques used in studies. Only by developing an acknowledged detection technique and stimulating a great deal of experimental and clinical data will the unified interpretation gradually be brought about.
With the promotion of precision medicine in NSCLC and the continued development of ALKis, more accurate auxiliary detection technology is the focus of liquid biopsy. Meanwhile, more laboratory evidence is needed to demonstrate the practicability of liquid biopsy in resistance surveillance. Real-time liquid biopsy is the trend in the application of resistance surveillance of targeted NSCLC therapy in ALK-positive patients. It will effectively guide patients with ALKi resistance to change therapy in time, prolong their lives and avoid the toxic side effects caused by ineffective treatment.
Acknowledgments
This work was supported by Nature Science Foundation of Guangdong Province, People’s Republic of China (2016A030313620).
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRLMillerKDJemalACancer statistics, 2017CA Cancer J Clin201767173028055103
- ChunhachaPChanvorachotePRoles of caveolin-1 on anoikis resistance in non small cell lung cancerInt J Physiol Pathophysiol Pharmacol20124314915523071872
- VázquezSCasalJAfonso AfonsoFJEGFR testing and clinical management of advanced NSCLC: a Galician Lung Cancer Group study (GGCP 048-10)Cancer Manag Res20168112026893581
- GuoWJLiuSHZhangXLThe coexpression of multi-immune inhibitory receptors on T lymphocytes in primary non-small-cell lung cancerDrug Des Devel Ther In press2017
- HamiltonGRathBDetection of circulating tumor cells in non-small cell lung cancerJ Thorac Dis201681024102827293809
- ScagliottiGVDe MarinisFRinaldiMItalian Lung Cancer ProjectPhase III randomized trial comparing three platinum-based doublets in advanced non-small-cell lung cancerJ Clin Oncol200220214285429112409326
- NestleUNiederCWalterKA palliative accelerated irradiation regimen for advanced non-small-cell lung cancer vs. conventionally fractionated 60 GY: results of a randomized equivalence studyInt J Radiat Oncol Biol Phys20004819510310924977
- YochumZASocinskiMABurnsTFParadoxical functions of ZEB1 in EGFR-mutant lung cancer: tumor suppressor and driver of therapeutic resistanceJ Thorac Dis2016811E1528E153128066651
- ValdespinoVValdespinoPMPotential of epigenetic therapies in the management of solid tumorsCancer Manag Res2015724125126346546
- FribouletLLiNKatayamaRThe ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancerCancer Discov20144666267324675041
- WangSCangSLiuDThird-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancerJ Hematol Oncol201693427071706
- WangSSongYYanFLiuDMechanisms of resistance to third-generation EGFR tyrosine kinase inhibitorsFront Med201610438338827770386
- IragavarapuCMustafaMAkinleyeANovel ALK inhibitors in clinical use and developmentJ Hematol Oncol201581725888090
- SodaMChoiYLEnomotoMIdentification of the transforming EML4-ALK fusion gene in non-small-cell lung cancerNature2007448715356156617625570
- ShawATYeapBYMino-KenudsonMClinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALKJ Clin Oncol200927264247425319667264
- TakeuchiKChoiYLTogashiYKIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancerClin Cancer Res20091593143314919383809
- SchaeferESBaikCProactive management strategies for potential gastrointestinal adverse reactions with ceritinib in patients with advanced ALK-positive non-small-cell lung cancerCancer Manag Res20168333827069372
- RikovaKGuoAZengQGlobal survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancerCell200713161190120318083107
- EttingerDSAkerleyWBorghaeiHNational comprehensive cancer networkNon-small cell lung cancerJ Natl Compr Canc Netw2012101236127123054877
- JoshiMRizviSMBelaniCPAfatinib for the treatment of metastatic non-small cell lung cancerCancer Manag Res20157758225733926
- SolomonBJMokTKimDWFirst-line crizotinib versus chemotherapy in ALK-positive lung cancerN Engl J Med2014371232167217725470694
- OuSHJännePABartlettCHClinical benefit of continuing ALK inhibition with crizotinib beyond initial disease progression in patients with advanced ALK-positive NSCLCAnn Oncol201425241542224478318
- OuSHAhnJSDe PetrisLAlectinib in crizotinib-refractory ALK-rearranged non-small-cell lung cancer: a phase II global studyJ Clin Oncol201634766166826598747
- ZouHYFribouletLKodackDPPF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical modelsCancer Cell2015281708126144315
- SolomonBJBauerTMFelipESafety and efficacy of lorlatinib (PF-06463922) from the dose-escalation component of a study in patients with advanced ALK+ or ROS1+ non-small cell lung cancer (NSCLC)J Clin Oncol2016349009
- ShawATFribouletLLeshchinerIResensitization to crizotinib by the lorlatinib ALK resistance mutation L1198FN Engl J Med20163741546126698910
- PagetSThe distribution of secondary growths in cancer of the breast. 1889Cancer Metastasis Rev198982981012673568
- RolfoCCastigliaMHongDLiquid biopsies in lung cancer: the new ambrosia of researchersBiochim Biophys Acta20141846253954625444714
- Quintás-CardamaAKantarjianHMCortesJEMechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemiaCancer Control200916212213119337198
- MokTKimDWWuYLFirst-line crizotinib versus pemetrexed-cisplatin or pemetrexed-carboplatin in patients (pts) with advanced ALK-positive non-squamous non-small cell lung cancer (NSCLC): results of a phase III study (PROFILE 1014)J Clin Oncol2014328002
- HeuckmannJMBalke-WantHMalchersFDifferential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variantsClin Cancer Res201218174682469022912387
- HeuckmannJMHölzelMSosMLALK mutations conferring differential resistance to structurally diverse ALK inhibitorsClin Cancer Res201117237394740121948233
- ShawATSolomonBKenudsonMMCrizotinib and testing for ALKJ Natl Compr Canc Netw20119121335134122157554
- KanedaHYoshidaTOkamotoIMolecularly targeted approaches herald a new era of non-small-cell lung cancer treatmentCancer Manag Res201359110123785245
- LindemanNICaglePTBeasleyMBMolecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular PathologyJ Thorac Oncol20138782385923552377
- CamidgeDRKonoSAFlaccoAOptimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatmentClin Cancer Res201016225581559021062932
- CondeEHernandezSPrietoMMartinezRLopez-RiosFProfile of Ventana ALK (D5F3) companion diagnostic assay for non-small-cell lung carcinomasExpert Rev Mol Diagn201616670771327031368
- JackmanDPaoWRielyGJClinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancerJ Clin Oncol201028235736019949011
- WuJSavoojiJLiuDSecond- and third-generation ALK inhibitors for non-small cell lung cancerJ Hematol Oncol201691926951079
- ToyokawaGSetoTUpdated evidence on the mechanisms of resistance to ALK inhibitors and strategies to overcome such resistance: clinical and preclinical dataOncol Res Treat201538629129826045026
- KatayamaRShawATKhanTMMechanisms of acquired crizotinib resistance in ALK-rearranged lung CancersSci Transl Med20124120120ra17
- ChoiYLSodaMYamashitaYALK Lung Cancer Study GroupEML4-ALK mutations in lung cancer that confer resistance to ALK inhibitorsN Engl J Med2010363181734173920979473
- SasakiTOkudaKZhengWThe neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancersCancer Res20107024100381004321030459
- KwakELBangYJCamidgeDRAnaplastic lymphoma kinase inhibition in non-small-cell lung cancerN Engl J Med2010363181693170320979469
- CroegaertKKolesarJMRole of anaplastic lymphoma kinase inhibition in the treatment of non-small-cell lung cancerAm J Health Syst Pharm201572171456146226294238
- DoebeleRCPillingABAisnerDLMechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancerClin Cancer Res20121851472148222235099
- GainorJFDardaeiLYodaSMolecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancerCancer Discov20166101118113327432227
- PetersSAdjeiAAGridelliCReckMKerrKFelipEESMO Guidelines Working GroupMetastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-upAnn Oncol201223Suppl 7vii56vii6422997455
- BolandJMJangJSLiJMET and EGFR mutations identified in ALK-rearranged pulmonary adenocarcinoma: molecular analysis of 25 ALK-positive casesJ Thorac Oncol20138557458123449277
- MosséYPWoodAMarisJMInhibition of ALK signaling for cancer therapyClin Cancer Res200915185609561419737948
- ChiarleRSimmonsWJCaiHStat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic targetNat Med200511662362915895073
- TakezawaKOkamotoINishioKJännePANakagawaKRole of ERK-BIM and STAT3-survivin signaling pathways in ALK inhibitor-induced apoptosis in EML4-ALK-positive lung cancerClin Cancer Res20111782140214821415216
- KatayamaRKhanTMBenesCTherapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALKProc Natl Acad Sci U S A2011108187535754021502504
- RenSWuCLiXZhaoCHouLZhouCIncidence of inconsistent driver mutations between multiple lung ground-glass nodules in patients with non-small cell lung cancerJ Clin Oncol20143215 Suppl11068
- ChaYJChoBCKimHRLeeHJShimHSA case of ALK-rearranged adenocarcinoma with small cell carcinoma-like transformation and resistance to crizotinibJ Thorac Oncol2016115e55e5826752677
- TakegawaNHayashiHIizukaNTransformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinibAnn Oncol2016275953955
- Ignatius OuSHAzadaMHsiangDJNext-generation sequencing reveals a Novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinibJ Thorac Oncol20149454955324736079
- YouLShouJDengDCrizotinib induces autophagy through inhibition of the STAT3 pathway in multiple lung cancer cell linesOncotarget2015637402684028226384345
- GuptaGPMassaguéJCancer metastasis: building a frameworkCell2006127467969517110329
- AllanALKeeneyMCirculating tumor cell analysis: technical and statistical considerations for application to the clinicJ Oncol2010201042621820049168
- TossAMuZFernandezSCristofanilliMCTC enumeration and characterization: moving toward personalized medicineAnn Transl Med201421110825489582
- TammingaMGroenHHHiltermannTJInvestigating CTCs in NSCLC-a reaction to the study of Jia-Wei Wan: a preliminary study on the relationship between circulating tumor cells count and clinical features in patients with non-small cell lung cancerJ Thorac Dis2016861032103627293811
- MengSTripathyDFrenkelEPCirculating tumor cells in patients with breast cancer dormancyClin Cancer Res200410248152816215623589
- LeongSMTanKMChuaHWSampling circulating tumor cells for clinical benefits: how frequent?J Hematol Oncol201587526108208
- ZhangZShiratsuchiHPalanisamyNNagrathSRamnathNExpanded circulating tumor cells from a patient with ALK-positive lung cancer present with EML4-ALK rearrangement along with resistance mutation and enable drug sensitivity testing: a case studyJ Thorac Oncol201712239740227507192
- AietaMFacchinettiADe FaveriSMonitoring and characterization of circulating tumor cells (CTCs) in a patient with EML4-ALK-positive non-small cell lung cancer (NSCLC)Clin Lung Cancer2016175e173e17727397482
- YangJJZhangXCSuJLung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylationClin Cancer Res20142051383139224443522
- NormantEPaezGWestKAThe Hsp90 inhibitor IPI-504 rapidly lowers EML4-ALK levels and induces tumor regression in ALK-driven NSCLC modelsOncogene201130222581258621258415
- GainorJFShawATEmerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancerJ Clin Oncol201331313987399624101047
- KuangYRogersAYeapBYNoninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancerClin Cancer Res20091582630263619351754
- SunWYuanXTianYNon-invasive approaches to monitor EGFR-TKI treatment in non-small-cell lung cancerJ Hematol Oncol201589526227959
- ShawJAStebbingJCirculating free DNA in the management of breast cancerAnn Transl Med201421325332979
- JahrSHentzeHEnglischSDNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cellsCancer Res20016141659166511245480
- DiehlFSchmidtKChotiMACirculating mutant DNA to assess tumor dynamicsNat Med200814998599018670422
- NewmanAMBratmanSVToJAn ultrasensitive method for quantitating circulating tumor DNA with broad patient coverageNat Med201420554855424705333
- JenkinsSYangJCRamalingamSSPlasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancerJ Thorac Oncol20171271061107028428148
- ChabonJJSimmonsADLovejoyAFCirculating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patientsNat Commun201671181527283993
- EscriuCFieldJKCirculating tumour DNA and resistance mechanisms during EGFR inhibitor therapy in lung cancerJ Thorac Dis2016892357235927746975
- WangYTianPWWangWYNoninvasive genotyping and monitoring of anaplastic lymphoma kinase (ALK) rearranged non-small cell lung cancer by capture-based next-generation sequencingOncotarget2016740652086521727564104
- CooperMRChimHChanHDurandCCeritinib: a new tyrosine kinase inhibitor for non-small-cell lung cancerAnn Pharmacother201549110711225258420
- OuSILeeTKYoungLDual occurrence of ALK G1202R solvent front mutation and small cell lung cancer transformation as resistance mechanisms to second generation ALK inhibitors without prior exposure to crizotinib. Pitfall of solely relying on liquid re-biopsy?Lung Cancer201710611011428285684
- GridelliCPetersSSgambatoACasaluceFAdjeiAACiardielloFALK inhibitors in the treatment of advanced NSCLCCancer Treat Rev201440230030623931927
- PunnooseEAAtwalSLiuWEvaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinibClin Cancer Res20121882391240122492982
- GevaSRoismanLCPeledNLiquid biopsy in the practice of neooncologyJ Thorac Dis2016810E1279E128127867607
- ZhouQYangJJChenZHSerial cfDNA assessment of response and resistance to EGFR-TKI for patients with EGFR-L858R mutant lung cancer from a prospective clinical trialJ Hematol Oncol2016918627619632
- RodríguezMSilvaJLópez-AlfonsoADifferent exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancerGenes Chromosomes Cancer201453971372424764226
- TavernaSGiallombardoMGil-BazoIExosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practiceOncotarget2016719287482876026919248
- ThéryCZitvogelLAmigorenaSExosomes: composition, biogenesis and functionNat Rev Immunol20022856957912154376
- MathivananSFahnerCJReidGESimpsonRJExoCarta 2012: database of exosomal proteins, RNA and lipidsNucleic Acids Res201240Database issueD1241D124421989406
- Demory BecklerMHigginbothamJNFranklinJLProteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRASMol Cell Proteomics201312234335523161513
- TaylorDDGercel-TaylorCMicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancerGynecol Oncol20081101132118589210
- KosakaNTakeshitaFYoshiokaYExosomal tumor-suppressive microRNAs as novel cancer therapy: “exocure” is another choice for cancer treatmentAdv Drug Deliv Rev201365337638222841506
- JaiswalRGongJSambasivamSMicroparticle-associated nucleic acids mediate trait dominance in cancerFASEB J201226142042921965597
- PigatiLYaddanapudiSCIyengarRSelective release of microRNA species from normal and malignant mammary epithelial cellsPLoS One2010510e1351520976003
- ChuangJCJonesPAEpigenetics and microRNAsPediatr Res2007615 Pt 224R29R
- LuJGetzGMiskaEAMicroRNA expression profiles classify human cancersNature2005435704383483815944708
- AnglicheauDMuthukumarTSuthanthiranMMicroRNAs: small RNAs with big effectsTransplantation201090210511220574417
- ShyuABWilkinsonMFvan HoofAMessenger RNA regulation: to translate or to degradeEMBO J200827347148118256698
- CalinGACroceCMMicroRNA signatures in human cancersNat Rev Cancer200661185786617060945
- GarzonRCalinGACroceCMMicroRNAs in cancerAnnu Rev Med20096016717919630570
- GalluzziLMorselliEVitaleImiR-181a and miR-630 regulate cisplatin-induced cancer cell deathCancer Res20107051793180320145152
- DacicSKellyLShuaiYNikiforovaMNmiRNA expression profiling of lung adenocarcinomas: correlation with mutational statusMod Pathol201023121577158220818338
- WuXXiaoHmiRNAs modulate the drug response of tumor cellsSci China C Life Sci200952979780119802736
- YanaiharaNCaplenNBowmanEUnique microRNA molecular profiles in lung cancer diagnosis and prognosisCancer Cell20069318919816530703
- EnfieldKSStewartGLPikorLAMicroRNA gene dosage alterations and drug response in lung cancerJ Biomed Biotechnol2011201147463221541180
- VlassovAVMagdalenoSSetterquistRConradRExosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentialsBiochim Biophys Acta20121820794094822503788
- NilssonRJKarachaliouNBerenguerJRearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancerOncotarget2016711066107526544515
- ChenSZhaoJCuiLLiuYUrinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIsClin Transl Oncol201719333234027468867
- LiYZhangYQiuFQiuZProteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLCElectrophoresis201132151976198321557262
- ZhangLXiaoHZhouHDevelopment of transcriptomic biomarker signature in human saliva to detect lung cancerCell Mol Life Sci201269193341335022689099
- WeiFYangJWongDTDetection of exosomal biomarker by electric field-induced release and measurement (EFIRM)Biosens Bioelectron20134411512123402739
- WeiFLinCCJoonANoninvasive saliva-based EGFR gene mutation detection in patients with lung cancerAm J Respir Crit Care Med2014190101117112625317990
- PuDLiangHWeiFEvaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: a pilot studyThorac Cancer20167442843627385985
- PaoWWangTYRielyGJKRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinibPLoS Med200521e1715696205
- MeloskyBTreatment algorithms for patients with metastatic non-small cell, non-squamous lung cancerFront Oncol2014425625325013
- YeTPanYWangRAnalysis of the molecular and clinicopathologic features of surgically resected lung adenocarcinoma in patients under 40 years oldJ Thorac Dis20146101396140225364516
- FaraceFMassardCVimondNA direct comparison of Cell-Search and ISET for circulating tumour-cell detection in patients with metastatic carcinomasBr J Cancer2011105684785321829190
- PaillerEOulhenMBorgetICirculating tumor cells with aberrant ALK copy number predict progression-free survival during crizotinib treatment in ALK-rearranged non-small cell lung cancer patientsCancer Res20177792222223028461563
- Zito MarinoFRoccoGMorabitoAA new look at the ALK gene in cancer: copy number gain and amplificationExpert Rev Anticancer Ther201616549350226943457
- BratmanSVNewmanAMAlizadehAADiehnMPotential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-SeqExpert Rev Mol Diagn201515671571925773944
- WangCWangAWeiFWongDTWTuMElectric field-induced disruption and releasing viable content from extracellular vesiclesMethods Mol Biol2017166036737628828672
- ReckMPopatSReinmuthNDe RuysscherDKerrKMPetersSESMO Guidelines Working GroupMetastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-upAnn Oncol201425 Suppl 3iii27iii3925115305
