Abstract
Hepatocellular carcinoma (HCC) is the third most common cause of death from cancer, with increasing prevalence worldwide. The mortality rate of HCC is similar to its incidence rate, which reflects its poor prognosis. At present, the diagnosis of HCC is still mostly dependent on invasive biopsy, imaging methods, and serum α-fetoprotein (AFP) testing. Because of the asymptomatic nature of early HCC, biopsy and imaging methods usually detect HCC at the middle–late stages. AFP has limited sensitivity and specificity, as many other nonmalignant liver diseases can also result in a very high serum level of AFP. Therefore, better biomarkers with higher sensitivity and specificity at earlier stages are greatly needed. Since metabolic reprogramming is an essential hallmark of cancer and the liver is the metabolic hub of living systems, it is useful to investigate HCC from a metabolic perspective. As a noninvasive and nondestructive approach, metabolomics provides holistic information on dynamically metabolic responses of living systems to both endogenous and exogenous factors. Therefore, it would be conducive to apply metabolomics in investigating HCC. In this review, we summarize recent metabolomic studies on HCC cellular, animal, and clinicopathologic models with attention to metabolomics as a biomarker in cancer diagnosis. Recent applications of metabolomics with respect to therapeutic and prognostic evaluation of HCC are also covered, with emphasis on the potential of treatment by drugs from natural products. In the last section, the current challenges and trends of future development of metabolomics on HCC are discussed. Overall, metabolomics provides us with novel insight into the diagnosis, prognosis, and therapeutic evaluation of HCC.
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of death from cancer, with increasing prevalence worldwide. The mortality rate of HCC is similar to its incidence rate, which reflects its poor prognosis.Citation1 HCC can be initiated by various factors, including genetic and environmental factors. The majority of HCC comes with a background of liver cirrhosis, primarily induced by two prominent risk factors: hepatitis B and C viruses (HBV and HCV) infection.Citation2 Chronic HBV infection consists of 52.3% and chronic HCV infection 20% of all HCC cases all over the world.Citation3 HBV is the leading etiologic factor for HCC in developing regions, such as western Africa and Southeast Asia, whereas HCV contributes to the most risk in developed countries. These two potent risk factors shape the geographical variation in HCC incidence, which in the developing world is higher than that in the developed world. Also, hereditary conditions, including genetic tyrosinemia, α1-antitrypsin deficiency, and hemochromatosis, can lead to HCC. An escalating amount of lifestyle-related risk factors, such as chronic alcohol abuse, tobacco smoke, betel-quid chewing, high aflatoxin intake, obesity, and diabetes, have also been linked with HCC.Citation4–Citation7 Invalid diagnosis and treatment of HCC often result in its high mortality rate, which poses a big threat to public health. Currently, the diagnosis of HCC is still mostly dependent on invasive biopsy, imaging methods, and serum α-fetoprotein (AFP) testing.Citation8 Because of early HCC asymptomatic nature, biopsy and imaging methods usually detect HCC at middle or late stages, for which there are no effectively therapeutic options. For the clinical diagnosis of HCC, serum AFP, a fetal serum glycoprotein, has been considered a golden biomarker. However, its sensitivity and specificity is very limited, because many other nonmalignant liver diseases, such as acute and chronic hepatitis and liver cirrhosis, can also result in a very high serum level of AFP that is similar to that detected in HCC.Citation9 Therefore, better biomarkers are greatly needed, which would improve clinical diagnosis and therapeutic treatment of HCC at earlier disease stages and ultimately result in lower mortality rates. An ideal biomarker should satisfy the following conditions: target molecules with high sensitivity and specificity; markers to be detectable via a noninvasive way, such as blood and urine; and measurement techniques should be cheap, reliable, and robust across an extensive range of populations.
As the newest “-omics” science, metabolomics is the systematic study of small-molecule metabolites in living systems, which is defined as the “metabolome” (metabolites with an atomic mass <1.5 kDa).Citation10 This approach offers holistic information on dynamically metabolic responses of living systems to both endogenous and exogenous factors. The target of metabolomics is to detect and identify global small-molecule metabolic profiles of complex biological matrices. Compared with other -omics technologies (namely genomics, transcriptomics, and proteomics), where modification of substrates commonly occurs, metabolomics can provide information on metabolites that are directly produced in response to endogenous and exogenous factors.Citation11 It provides potential biomarkers for the diagnosis and monitoring of complex diseases and response to therapeutic intervention.Citation12,Citation13 It has been widely applied in the domains of disease diagnosis, therapeutic monitoring, and pharmacodynamic evaluation.Citation14–Citation16 Cell metabolism plays an important role in cancer, and metabolic reprogramming is an essential hallmark of cancer.Citation17 Cancer cells have been reported to own a significantly unique metabolic phenotype to support their high proliferation rates, which is highlighted by high glycolytic rates, enriched phospholipid turnover, low mitochondrial activity, and decreased bioenergetic expenditure.Citation18 Cancer metabolism has become a “hot spot” and is gaining momentum for better mechanistic research of tumorigenesis. Therefore, it is useful to investigate cancer from a metabolic perspective.
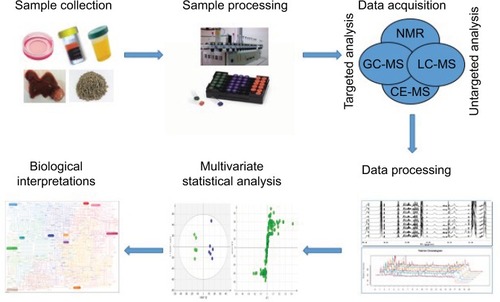
As the metabolic hub of the human body with various metabolic functions, the liver is able to mediate the expression levels of numerous metabolites. It is well accepted that metabolic-profile variations occur prior to imaging diagnosis in HCC patients. Owing to this, HCC has become a disease model of great interest from a metabolic perspective.Citation19,Citation20 More specifically, metabolomics is a noninvasive and nondestructive analysis. Therefore, it would be conducive to apply metabolomics to understand the complex pathophysiology, discover new biomarkers, and evaluate new therapeutic drug targets of HCC. There has been an increasing number of metabolomic studies on HCC over the last few decades.Citation21,Citation22 The typical processing flow of metabolomics studies on HCC is shown in . Firstly, samples for HCC are collected. There are different kinds of samples used for HCC, including in vitro HCC cells, in vivo plasma, serum, urine, feces, and liver tumor and nontumor tissue. Collected samples are preprocessed and tested by different approaches. There are two main analytical platforms used in metabolomic studies on HCC: nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). The latter is usually equipped with different separation instruments, including liquid chromatography (LC), gas chromatography (GC), and capillary electrophoresis (CE). Each platform has its own strengths and weaknesses. After acquisition, the data obtained are processed and analyzed to find metabolites changed majorly by multivariate statistical analysis, such as principal-component analysis and orthogonal projection to latent structure-discriminant analysis. Lastly, according to databases and related literature, the metabolites involved and corresponding pathways are elucidated and the specific biomarkers identified.
Figure 1 Typical processing flow of metabolomics in HCC.
Notes: Proposed standards for metabolomics on HCC are presented in this schematic view. The first step is to collect the samples. Then, the collected samples are preprocessed and tested by different approaches. After acquisition, obtained data are processed and analyzed by multivariate statistical analysis. Lastly, the underlying biological interpretations are elucidated.
Abbreviations: CE, capillary electrophoresis; GC, gas chromatography; HCC, hepatocellular carcinoma; LC, liquid chromatography; MS, mass spectrometry; NMR, nuclear magnetic resonance.
For this review, we retrieved data from metabolomic studies on HCC in the prior 7 years from the databases PubMed and Google Scholar using the five keywords “hepatocellular carcinoma”, “metabolomics”, “biomarker”, “diagnosis”, and “therapy”. First, we summarized the recent metabolomic studies on HCC cellular, animal, and clinicopathologic models with attention to its use as a biomarker in cancer diagnosis. Subsequently, recent applications of metabolomics with respect to therapeutic and prognostic evaluation of HCC were also focused upon, with emphasis on the potential of treatment by drugs from natural products. In the last section, current challenges of and trends in future development of metabolomics with regard to HCC are discussed. Overall, metabolomics provides us a novel insight into diagnosis, prognosis, and therapeutic evaluation of HCC.
Metabolomics in HCC-model research
In this section, we summarize recent metabolomic studies on HCC cellular, animal, and clinicopathologic models with attention to their use as biomarkers in cancer diagnosis. depicts detailed information on four metabolomic studies on HCC cells and nine HCC animal models published recently, and includes detailed information on 37 metabolomic studies of human HCC samples. Overall, these studies illustrated the majorly altered metabolites and corresponding metabolic pathways involved in stepwise hepatocarcinogenesis, suggesting that metabolomics could be a promising strategy of biomarker discovery for the early diagnosis of HCC.
Table 1 Summary of recent metabolomics studies on HCC cellular and animal models
Table 2 Summary of recent metabolomic studies on HCC clinicopathologic models
Cell pathology model research
HBV infection involves intricate interactions between the virus and host cells that can induce HCC. It has been reported that there was a correlation between HBV infection and metabolic alterations in host cells.Citation23,Citation24 To study the underlying mechanisms of metabolic alterations caused by HBV infection, NMR-based metabolomics was used to investigate the metabolic features of HepG2.2.15 cells derived from HepG2 with stable expression and replication of HBV, which is a commonly used cell model to study HBV infection.Citation25 The results indicated that HBV infection contributed to HCC by upregulation of the glutamine-fructose-6-phosphate amidotransferase 1 (GFAT1)-activated hexosamine biosynthesis and choline kinase alpha (CHKA)-activated phosphatidylcholine biosynthesis. Hepatitis B virus X protein (HBx), a multifunctional oncoprotein, was reported to be associated with HBV replication, DNA repair, cell-cycle progression, transcriptional regulation, and to play an essential role in HBV-related HCC.Citation26 Yue et al used an NMR-based metabolomic approach to systematically investigate the effects of this specific protein on hepatocarcinogenesis.Citation27 They found that HBx disrupted the metabolism of glucose, lipids, and amino acids, especially nucleic acids. Further investigation of the effects of HBx on nucleic acid metabolism in gene-expression profiles showed that 29 genes correlating with DNA damage and repair in HBx-expressing HepG2 cells were differentially expressed. Together, their results revealed that HBx initially caused DNA damage and then perturbed nucleic acid metabolism, which in turn blocked DNA repair and led to HCC. E4F1, a cellular target of the E1A adenoviral oncoprotein, was reported to interact with HBx. Based on combined analyses of 1H-NMR metabolomics and molecular biology technologies, Dai et al confirmed that E4F1 may contribute to the proliferation of HBV-infected HCC cells by neutralizing the capacity of HBx to activate a p53-dependent metabolic and growth arrest phenotype.Citation28 HBc, encoded by the HBV genome, was also reported to play an essential role in HBV-related HCC. Xie et al combined analyses of proteomics and metabolomics to investigate the function of HBc in HBV-related HCC.Citation29 They found that HBc upregulated glycolysis and amino acid metabolism, and the enriched recruitment and enrichment of Mlx by HBc in the nucleus were linked to glycolysis pathways. PNPLA3 is a gene regulating both acylglycerol O-acyltransferase and triacylglycerol lipase activities. To investigate its role in HCC, Min et al performed GC-MS and LC-MS metabolic profiling of Huh7 cells with PNPLA3 siRNA silencing and overexpression.Citation30 Silencing of PNPLA3 was revealed to reduce amino acid metabolism and elevate levels of myoinositol, cysteine sulfinic acid, polyunsaturated fatty acids, lysolipids, and sphingolipids. Overexpression of PNPLA3 mirrored metabolic changes in the opposite direction. Taken together, their results revealed a central role of PNPLA3 in the regulation of liver metabolism, besides its traditional role in the remodeling of triacylglycerol.
Animal pathologic model research
Different HCC animal models have been established to investigate the pathogenesis of HCC and identify specific biomarkers. Based on these models, a large number of metabolomic studies have been performed. Chemicals can lead to tumorigenesis in the liver of rodents, and diethylnitrosamine (DEN) has been reported to be the most commonly used hepatocarcinogen in rodents. It has been reported that the genetic and histologic signatures of the DEN-induced HCC rodent model are similar to those of human HCC.Citation31 Several metabolomic studies have been conducted to compare the metabolic profiles of control and HCC using DEN-induced rodents. For example, Hong et al used DEN-induced C57BL/6J model to investigate the role of estrogen-related receptor α (ERRα) in HCC.Citation32 LC-MS/MS-based metabolomic analysis revealed that in response to DEN, the loss of ERRα led to hepatocyte necrosis via apoptosis, which induced HCC through independent but synergistic mechanisms of hepatocytes and Kupffer cells. Tan et al used LC-MS-based metabolomics to identify potential biomarkers from the serum of a DEN-induced rat HCC model.Citation33 Three metabolites – lysophosphoethanolamine 16:0, lysophosphatidylcholine (LPC) 22:5, and taurocholic acid – were identified as biomarkers and effective in discriminating HCC patients better than AFP in sensitivity and specificity. Similar samples were also analyzed via CE–time of flight (TOF)–MS-based metabolomics.Citation34 To improve the detection of early risk of HCC, Huang et al proposed a new strategy for analysis of time-series data based on dynamic networks (ATSD-DN) using the DEN-induced rat HCC model.Citation35 LC-MS-based metabolomic analysis of serum identified a ratio of lysophosphatidylcholine (LPC) 18:1/free fatty acid 20:5 as a biomarker of HCC. The better performance of ATSD-DN suggested its potential to represent time-series changes well and effectively extract early-warning information. Also, the DEN-induced rat HCC model can develop lung metastatic nodules and has also been used to investigate the metastasis of HCC, which contributes to the poor prognosis and high mortality rate of HCC. Li et al used a DEN-induced rat HCC with lung metastasis (HLM) model to study serum and urine metabolic profiles via GC/TOF-MS metabolomics.Citation36 They found that glutamate metabolism and glycolysis were increased and the tricarboxylic acid (TCA) cycle decreased in both HCC and HLM. Especially, the metabolism of glucuronic acid, amino acids, and nucleic acids were increased only in HLM, which revealed potential biomarkers for HCC invasion and metastasis. Wang et al also used this model to study tumor-tissue metabolic profiles via 1H-NMR metabolomics.Citation37
The transgenic mouse model is also used to investigate the pathogenesis of HCC. Teng et al used an HBx transgenic mouse model to study HBV-associated HCC pathogenesis.Citation38 GC-MS-based metabolic profiles of serum and liver revealed that lipid (fatty acids, triglycerides, and cholesterol) profiles changed significantly during the development of HBx tumorigenesis, supporting the view that metabolic syndrome is essential in HBV tumorigenesis. Fan et al used a Ras-Tg mice model to study Hras12V oncogene-associated HCC pathogenesis.Citation39 The combined analysis of transcriptomics and GC-TOF-MS-based metabolomics revealed that Hras12V induced perturbations of glycolysis, the pentose phosphate pathway, the TCA cycle, lipid metabolism, bile-acid synthesis, and redox homeostasis. These altered metabolic pathways may be essential to HCC. In addition, an HCC-xenograft model has also been used. Alcohol abuse is one of the main causes of liver injury that can promote HCC. To investigate the underlying pathogenesis of HCC from liver injury, an HCC-xenograft model was built by subcutaneously inoculating HepG2 cells into nude mice.Citation40 Ultrahigh-performance LC (UHPLC)/quadrupole TOF (QTOF)-MS-based metabolic profiles of serum revealed metabolic alterations of LPCs in liver injury and HCC, suggesting that the serum profile of LPC may be a biomarker for liver injury and HCC.
Clinicopathologic model research
Apart from research on HCC cellular and animal pathologic models, there have been a larger number of metabolomic studies on HCC clinicopathologic models. Since blood, urine, and feces can be easily obtained from humans without any invasion, they are commonly used in human metabolomic studies. A number of metabolomic studies of blood and urine to distinguish HCC from healthy controls have been reported. Assi et al performed 1H-NMR-based metabolomics on prediagnostic serum from 114 first-incident, primary HCC cases and 222 matched controls identified from among the participants of a large European prospective cohort.Citation41,Citation42 They found that the changed metabolites in the HCC group were positively related to HCC risk. LC-MS-based metabolomic studies have also been done.Citation43–Citation47 Chen et al performed LC-MS-based metabolomics on serum from 41 HCC patients and 38 healthy controls.Citation43 They identified the serum 1-methyladenosine as a characteristic metabolite for HCC, and a combination of 1-methyladenosine and AFP showed significantly improved sensitivity. Chen et al conducted UHPLC/triple – quadrupole MS-based metabolomics on serum from 29 HCC patients and 30 age-matched healthy controls.Citation44 Lower LPC, medium-chain acylcarnitines, branched-chain amino acids and enriched long-chain acylcarnitines, and aromatic amino acids were observed in HCC patients. Liang et al conducted LC-QTOF-MS-based metabolomics on urine from 25 HCC patients and 12 matched healthy controls.Citation46 The citric acid cycle, bile-acid biosynthesis, urea-cycle metabolism, and tryptophan metabolism were significantly changed in the HCC group. In addition, a study on serum and urine metabolic profiles from 82 HCC patients, 24 benign liver tumor patients, and 71 healthy controls was conducted by a combination of GC-TOF-MS and UHPLC-QTOF-MS metabolomics:Citation48 43 serum metabolites and 31 urine metabolites were identified in HCC patients involving the metabolism of glycolysis, free fatty acids, bile acids, methionine, and the urea cycle. The identified biomarkers differentiated HCC patients with AFP levels lower than 20 ng/mL from healthy controls with 100% accuracy.
The early and accurate discrimination of HCC from other high-risk liver diseases, such as HBV, HCV, and liver cirrhosis, can improve the prognosis of HCC patients. As the majority of HCC comes with a background of liver cirrhosis, many studies have attempted to discriminate the serum, plasma, urine, or feces metabolic profiles of HCC from liver cirrhosis.Citation49–Citation64 For example, Nahon et al performed 1H-NMR-based metabolomics on serum from 93 liver cirrhosis, 28 small HCC, and 33 large HCC samples.Citation49 Compared with cirrhosis, levels of glutamate, acetate, and N-acetyl glycoproteins were increased and levels of lipids and glutamine largely decreased in HCC. A combination of NMR and LC-MS-based metabolomics was applied to study serum metabolic profiles from 43 HCC patients, 42 liver cirrhosis patients, and 18 healthy volunteers.Citation52 The results revealed that HCC induced disturbances of the citrate cycle, amino acid catabolism, fatty-acid oxidation, phospholipid metabolism, synthesis of ketone bodies, bile-acid metabolism, and sphingolipid metabolism. Sanabria et al conducted GC/LC-MS-based metabolomic analyses on plasma from healthy subjects (n=20), patients with end-stage liver disease (n=99), and patients after liver transplantation (n=7). The results showed glutathione species may define liver disease severity and serve as surrogates for the early detection of HCC in patients with established cirrhosis.Citation57 Shao et al conducted QTrap LC-MS-based metabolomics on urine from 27 liver cirrhosis subjects, 33 HCC subjects, and 26 healthy individuals,Citation59 identifying hydantoin-5-propionic acid and butyrylcarnitine (carnitine C4:0) as combinational biomarkers to distinguish HCC from liver cirrhosis. Cao et al performed UHPLC/QTOF-MS-based metabolomics on feces from 23 healthy individuals, 22 with liver cirrhosis, and 23 with HCC.Citation63 Compared with healthy controls, LPC was significantly increased and bile acids and bile pigments significantly decreased in liver cirrhosis and HCC, which were identified as potential biomarkers. Since HBV/HCV infection is the prominent inducer of HCC, there have been several metabolomic investigations on the metabolic differences between hepatitis patients and HCC patients.Citation65–Citation74 Bowers et al performed HPLC-MS-based metabolomics on serum from 37 HCC patients and 21 HCV patients.Citation67 Levels of cholylglycine, xanthine, uric acid, dioleoylphosphatidylcholine, arachidonyl lysolecithin, 3-hydroxycapric acid, and D-leucic acid were found to be altered in HCC, and may be potential biomarkers to distinguish HCC from HCV. Wei et al distinguished HCC from HCV by three metabolites (choline, valine, and creatinine) with 1H-NMR-based serum metabolomics.Citation68 Gao et al performed GC-TOF-MS-based metabolomics on serum from 49 HBV patients, 52 liver cirrhosis patients, 39 HCC patients, and 61 healthy subjects (normal controls [NC]).Citation70 β-Glutamate and asparagine for HCC versus liver cirrhosis, palmitic acid for liver cirrhosis versus HBV, and 5-methoxytryptamine, malic acid, and phenylalanine for HBV versus NC were selected as potential liver disease-specific biomarkers. Gong et al used a combination of untargeted metabolomics and targeted eicosanoid analysis on serum from 49 HBV-cirrhosis patients, 51 HCC patients, and 39 NC. The biomarkers identified in this study showed high potential to differentiate HCC from NC and HBV-cirrhosis patients.Citation71
Besides the commonly used blood and urine samples in clinical metabolomic studies, there have also been metabolomic studies based on human tissue.Citation75–Citation80 The liver is the most essential metabolic organ, and liver extracts can provide complex metabolic information. Additionally, integration of metabolomics with other -omics technologies can be achieved when the sample used is tissue. For example, Budhu et al performed an integration of gene-expression technology, LC-MS, and GC-MS-based metabolomics on paired tumor and nontumor tissues from 30 HCC patients:Citation77 169 genes and 28 metabolites were identified to be related to HCC. Specifically, monounsaturated palmitic acid, a product of stearoyl-CoA-desaturase, was elevated in aggressive HCC and confirmed to increase the invasion and migration of HCC cells in vitro. Beyoğlu et al conducted an integration of transcriptomics and GC-MS-based metabolomics on 31 pairs of HCC tumors and corresponding nontumor liver tissues.Citation78 The metabolomic results showed that levels of glucose, malate, myoinositol, alanine, linoleic acid, and glycerol 2- and 3-phosphate were decreased in HCC. Transcriptomics classified HCC in G1–G6 subgroups, and suggested that the high serum level of AFP in G1 was connected to the overexpression of lipid catabolic enzymes. Altogether, HCC highlighted metabolic remodeling from mitochondrial oxidation to aerobic glycolysis. In addition, the combination of metabolomics with other -omics technologies could also be used to study the differences between HCC and intrahepatic cholangiocarcinoma.Citation79
Metabolomics in treatment evaluation
Besides metabolomic studies on HCC, recent applications of metabolomics with respect to therapeutic and prognostic evaluation of HCC have also focused on the potential of treatment by drugs from natural products (). Metabolomics provides comprehensive metabolic profiles of living systems in response to drug treatment or surgery, which further helps us to better understand the underlying mechanism of anti-HCC agents and predict the risk of tumor recurrence.
Table 3 Summary of recent metabolomic applications with respect to efficacy of anti-HCC agents and prognostic evaluation
Metabolomics in anti-HCC-agent research
Chemotherapy remains one of the main approaches for the treatment of HCC. Specifically, metabolomics can be used to study the effects of cotreatment of currently approved drugs. NMR-based metabolomics was applied to investigate the effects of belinostat and bortezomib cotreatment on HCC.Citation81 The cotreatment increased amino acids and induced oxidative stress, displaying synergistic antiproliferative properties. A similar synergistic anti-HCC effect was also found in sorafenib–everolimus combination therapy using 2-D high-resolution magic-angle spinning 1H-NMR metabolomics.Citation82 So far, most chemotherapeutic drugs have not been effective for HCC, due to drug resistance and serious side effects.Citation83 Therefore, it is urgently necessary to develop new anti-HCC agents. Due to their multiple biological activities and low toxicity, natural products obtained from Chinese medicine have been explored to improve HCC prognosis and survival rate. The in vitro application of metabolomics for efficacy evaluation of anti-HCC agents from natural products has been shown to be predictive of treatment efficacy. Geranylgeranic acid (GGA), an acyclic diterpenoid, is found in medicinal herbs, such as turmeric. It has been reported to induce the death of HCC Huh7 cells. Iwao et al used UHPLC/TOF-MS-based metabolomics to study the underlying mechanism of its anti-HCC effect.Citation84 Their results showed that GGA could increase fructose 6-phosphate and spermine while decreasing fructose 1,6-diphosphate and spermidine, suggesting that GGA may shift Huh7 cells from aerobic glycolysis to mitochondrial respiration via upregulating TIGAR and SCO2 protein levels. Dehydroepiandrosterone (DHEA), a steroid secreted by the brain, gastrointestinal tract, adrenal cortex and gonads, is reported to have antiproliferative properties. A rapid-resolution LC-TOF-MS metabolomic approach revealed that DHEA treatment caused changes in metabolism of glutathione, lipids, S-adenosylmethionine, and lysine, suggesting that the mitochondrial dysfunction caused by decreased S-adenosylmethionine production and cardiolipin depletion underlay the antiproliferative effect of DHEA.Citation85 Ikuta et al used GC-MS metabolomics to evaluate the effect of R-α-lipoic acid (RLA) on rat HCC H4IIEC3 cells.Citation86 The results showed RLA treatment inhibited gluconeogenesis via suppressing Thr–Gly–Ser pathways, glycolysis, and lactic acid production, providing the mechanism on how RLA induces apoptosis in HCC cells. Acyclic retinoid (ACR), a synthetic retinoid, was shown to prevent HCC but not normal hepatic cells. Qin et al applied NMR and CE-TOF-MS-based metabolomics to investigate the molecular basis of its selective anti-HCC effect.Citation87 It was found that ACR treatment suppressed adenosine triphosphate, suggesting that ACR played a selective anti-HCC effect via suppression of the enhanced energy metabolism of HCC but not normal hepatic cells. Noguchi et al used GC-MS-based metabolomics to investigate the effects of cotreatment of palmitate (PA) and oleate (OA) on HCC.Citation88 They found that metabolite levels for TCA-cycle intermediates, pentose phosphate-pathway intermediates, and energy-storage metabolites changed between PA treatment alone and PA-OA cotreatment. This demonstrated that abnormal pentose phosphate-pathway fluxes and increased adenosine levels might contribute to differences between PA treatment alone and PA-OA cotreatment.
Metabolomics has also been applied to evaluate the efficacy of anti-HCC natural products in vivo. For example, 6,7-dimethoxy-1,2,3,4-tetrahydro-isoquinoline-3-carboxylic acid is an isoquinoline alkaloid extracted from Mucuna pruriens seeds and reported to have antiproliferative action. Kumar et al used 1H-NMR-based metabolomics of serum from HCC rats to investigate the underlying mechanism.Citation89 The results showed that 6,7-dimethoxy-1,2,3,4-tetrahydro-isoquinoline-3-carboxylic acid treatment modulated fatty acids, low-density lipoproteins, acetoacetate, choline, lysine, leucine, isoleucine, tyrosine, pyruvate, and creatine to normal levels, and amelioration of these markers may be linked to repair of inflammation damage, improvement in energy metabolism, and reconstruction of cell-membrane injury. Li et al applied UHPLC-QTOF-MS-based metabolomics of urine samples from mice bearing H22 cells to investigate the anti-HCC effect of hispidulin, which is found widely in Chinese herbs.Citation90 Physapubenolide is a withanolide extracted from Physalis pubescens. Ma et al applied GC-MS-based metabolomics to explore its anti-HCC mechanism.Citation91 They found that physapubenolide significantly decreased lactate production and suppressed glycolysis via the Akt–p53 pathway. Similar anti-HCC effects of ACR were also explored via CE-TOF-MS- and LC-TOF-MS-based metabolomics of liver tissue samples from the DEN-induced HCC mouse model.Citation92 Metabolomics can also be applied to investigate the anti-HCC effect of traditional Chinese medicine formulae. For example, shuihonghuazi formula (SHHZF) is a traditional Chinese medicine made from four herbs, namely Semen Coicis, Fructus Polygoni Orientalis, Imperatae Rhizoma, and Ophicalcitum. SHHZF has been applied to treat HCC clinically, and its underlying mechanism was investigated in liver tissue from a DEN-induced HCC rat model via HPLC/electrospray ionization–TOF-MS-based metabolomics.Citation93 It was observed that SHHZF inhibited abnormal fatty-acid and bile-acid metabolism, and its anti-HCC property was achieved via regulating the activities of lysophospholipase D, MTHFR, and PEMT.
Metabolomics in prognostic evaluation
Surgery is the common curative approach for HCC patients, yet predicting the risk of tumor recurrence after surgery is difficult. Prognosis estimation in HCC patients is important, which can offer essential information on diagnosis and thus indicate treatment. Metabolomics is also applied to evaluate the prognosis of HCC. For example, Zhou et al used LC-MS-based metabolomics of plasma to predict early postoperative recurrence among HCC patients.Citation94 They collected plasma samples from 22 early-recurrent and 18 late-recurrent HCC patients. Compared with the late-recurrent HCC group, fatty-acid and bile-acid steroids in the early-recurrent HCC group were found to change greatly. Specifically, decreased levels of linolenic acid, docosahexaenoic acid, and polyunsaturated eicosapentaenoic acid were the specific biomarkers for early recurrence. Ye et al utilized GC-TOF-MS-based metabolomics to investigate the complex physiopathological regulations of HCC after surgical resection.Citation95 They collected urinary samples from 19 pairs of matched preoperative and postoperative HCC patients and 20 healthy volunteers. Their results showed that metabolism of the TCA cycle, amino acids, nucleosides, and gut flora significantly changed after surgical resection, and five metabolites (acotinic acid, phenylalanine, ethanolamine, ribose, and lactic acid) were identified as biomarkers to predict early recurrence. Goossens et al applied 1H-NMR-based metabolomics to investigate the risk of tumor recurrence in HCC patients.Citation96 They collected serum samples from 120 HCC patients before and after radiofrequency ablation. Their results revealed clear differences relying on whether HCC had a viral or nonviral etiology before radiofrequency ablation. Chen et al also applied 1H-NMR-based metabolomics to discriminate plasma metabolic profiles of HCC patients from different therapeutic backgrounds, such as transcatheter arterial chemoembolization or surgical treatment. They found that transcatheter arterial chemoembolization or surgical treatment did not immediately evoke apparent amelioration in the metabolic profiles of HCC patients.Citation97
Current challenges and future trends
Altogether, though great achievements have been made in the field of HCC-biomarker discovery and therapy evaluation by metabolomics, this discipline is still in its infancy, and much exciting work remains to be done.
Current challenges
Although metabolomics has become a hot spot in the scientific community and better understanding of the complex pathophysiology of HCC has been achieved, this discipline still lags behind other -omics technologies to a great extent.Citation98 The current challenges of metabolomics are primarily embodied in the following aspects. First, the most common challenge for metabolomics are its technical limitations. There are about 2,000 metabolites in living systems, and the number of metabolites detected by the current instrumentation is only in the hundreds. Although the development of analytical instrumentation has made much progress, most metabolites cannot be detected due to their dramatic concentration range and great complexity, and these undetectable metabolites may also play an important role in living systems. Second, as a high-throughput pattern-recognition method, metabolomics is based on the analysis of big data. Therefore, studies with small and inadequate samples would cause false-negative results. Third, although most recent studies have offered us detailed information, such as demographic data for human subjects, there is still a lack of consistency on how the samples were selected and classified. For example, liver compensation and the treatment status of human subjects have often been inconsistently reported among different metabolomic studies on HCC. Fourth, a great challenge exists in data analysis of metabolomics, which is the most time-consuming step of metabolomic studies. The limited database and analysis tools may lead to false-negative or false-positive results. Last but not least, a key challenge in metabolomics studies on HCC is bias among the researchers, which is an obstacle to bring metabolomics into clinical application. For example, very different biomarkers can be identified for the same cancer, due to different sample processing, different analytical platforms used, and different data processing.
Future trends
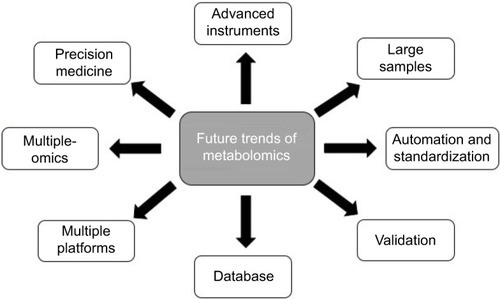
Further trends in metabolomic studies on HCC are as follows (). In order to detect as many metabolites as possible, efforts should be made to advance the analysis platforms and computational methods in metabolomic studies. It is anticipated that future metabolomic studies will involve larger epidemiological studies that cover analysis of thousands of samples instead of the presently small sample sizes.Citation99 A recently emerging trend in metabolomics is automation due to the need for quick and accurate quantification in the clinic that not only enlarges throughput but also elevates the reproducibility and reliability among different laboratories.Citation100 Besides, standardization of metabolomic protocols, including sample collecting, handling, storing, techniques, hardware, and data processing, can also improve the reproducibility and reliability of metabolomics.Citation101 A greatly recommended step of metabolomics is to validate metabolomic results. All the key metabolites obtained from metabolomic analysis should be validated to eliminate any unreliable findings by use of an additional set of samples. In the metabolomic studies on HCC we reviewed, only about 20% of publications had their findings validate. Therefore, it is a trend to validate key metabolites obtained by metabolomic analysis for both early HCC diagnosis and therapy evaluation.Citation102 In order to gain a better processing and biological interpretation of metabolomic data, a more comprehensive database will be developed based on global body function, rather than organ-related function. It is widely accepted that a single analytical instrument can only identify a limited number of detectable metabolites. Therefore, an integration of different analytical instruments will be a trend to extend the number of detectable metabolites in biological samples and offer us more information.Citation103 Most of the recent metabolomic studies on HCC have been conducted only at the metabolic level and lacked integrated information on other levels, such as DNA, RNA, and proteins. Since no single technology can provide us the entire spectrum of the HCC phenotype, integration of metabolomic results with markers obtained from the upstream -omics technologies is a new trend to increase our understanding of the pathological progression of HCC, as well as responses to therapy.Citation104 The concept of “precision medicine” (also called personalized medicine) in cancer health care is gradually gaining interest, of which the goal is to use advanced technologies to customize an individual’s medical treatment based on their own biomarker profiles. Given that metabolomics is relatively inexpensive and provides valuable biomarkers in disease diagnosis and therapy monitoring, there can be little doubt that it will be useful in precision medicine to guide clinical strategies from personal disease diagnosis and monitoring to selecting a suitable therapy and finally to following patient prognosis in the future.Citation105 Although the consideration of these challenges and future trends will not be satisfied at present or in the near future, it is essential that they are highly recognized and being discussed on a larger scale.
Conclusion
In summary, metabolomics is a burgeoning science with a combination of analytical chemistry, biochemistry, bioinformatics, and medicine, which is well suited to studies of HCC. It can be applied not only to identify the sensitive and specific biomarkers of HCC in a noninvasive and nondestructive way at the early stage but also to evaluate the efficacy of treatment and prognosis. Valuable information regarding HCC diagnosis, prognosis, and therapy can be obtained from these metabolomic studies of HCC. Collectively, metabolomics provides us new insights into the diagnosis, prognosis, and therapeutic evaluation of HCC. With the coming era of precision medicine, metabolomics will undoubtedly play an active role in selecting a suitable therapy tailored to an individual patient in the future. Although technological advances have brought metabolomics into the spotlight, metabolomics is still in its infancy, and much exciting work remains to be done.
Acknowledgments
The study was financially supported by grants from the research council of the University of Hong Kong (project codes 104004092, 104003919, 104004460, 104004745, and 104004746), the Research Grants Committee (RGC) of Hong Kong, HKSAR (project codes 766211, 17152116), Wong’s Donation for Modern Oncology of Chinese Medicine (project code 200006276), and the Shenzhen Basic Research Program (project code JCYJ20140903112959964).
Disclosure
The authors report no conflicts of interest in this work.
References
- PatelMShariffMILadepNGHepatocellular carcinoma: diagnostics and screeningJ Eval Clin Pract201218233534221114800
- OhataKHamasakiKToriyamaKIshikawaHNakaoKEguchiKHigh viral load is a risk factor for hepatocellular carcinoma in patients with chronic hepatitis B virus infectionJ Gastroenterol Hepatol200419667067515151623
- BoschFXRibesJDiazMCleriesRPrimary liver cancer: worldwide incidence and trendsGastroenterology20041275 Suppl 1S5S1615508102
- MorganTRMandayamSJamalMMAlcohol and hepatocellular carcinomaGastroenterology20041275 Suppl 1S87S9615508108
- TsaiJFChuangLYJengJEBetel quid chewing as a risk factor for hepatocellular carcinoma: a case-control studyBr J Cancer200184570971311237396
- GroopmanJDSchollPWangJSEpidemiology of human aflatoxin exposures and their relationship to liver cancerProg Clin Biol Res19963952112228895991
- RegimbeauJMColombatMMognolPObesity and diabetes as a risk factor for hepatocellular carcinomaLiver Transpl2004102 Suppl 1S69S73
- JustesenPFengerCHøilund-CarlsenPFHepatocellulært karcinom og andre levertumorer [Hepatocellular carcinoma and other liver tumors]Ugeskr Laeger2006168191876 Danish16756814
- TaketaKα-Fetoprotein: reevaluation in hepatologyHepatology1990126142014321701754
- WishartDSTzurDKnoxCHMDB: the Human Metabolome DatabaseNucleic Acids Res200735DatabaseD521D52617202168
- GantiSWeissRHUrine metabolomics for kidney cancer detection and biomarker discoveryUrol Oncol201129555155721930086
- ZhangASunHWangPHanYWangXRecent and potential developments of biofluid analyses in metabolomicsJ Proteomics20127541079108822079244
- CatchpoleGSBeckmannMEnotDPHierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato cropsProc Natl Acad Sci U S A200510240144581446216186495
- ArmitageEGSouthamADMonitoring cancer prognosis, diagnosis and treatment efficacy using metabolomics and lipidomicsMetabolomics20161214627616976
- Kaddurah-DaoukRWeinshilboumRMetabolomic signatures for drug response phenotypes: pharmacometabolomics enables precision medicineClin Pharmacol Ther2015981717525871646
- LiuCAlessandroAXiaYMetabolomic approach in probing drug candidatesCurr Top Med Chem201717151741174927848895
- WardPSThompsonCBMetabolic reprogramming: a cancer hallmark even Warburg did not anticipateCancer Cell201221329730822439925
- SerkovaNJSpratlinJLEckhardtSGNMR-based metabolomics: translational application and treatment of cancerCurr Opin Mol Ther20079657258518041668
- WangXZhangASunHPower of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinomaHepatology20135752072207723150189
- ChaiteerakijRAddissieBDRobertsLRUpdate on biomarkers of hepatocellular carcinomaClin Gastroenterol Hepatol201513223724524275343
- KimhoferTFyeHTaylor-RobinsonSThurszMHolmesEProteomic and metabonomic biomarkers for hepatocellular carcinoma: a comprehensive reviewBr J Cancer201511271141115625826224
- FitianAICabreraRDisease monitoring of hepatocellular carcinoma through metabolomicsWorld J Hepatol20179111728105254
- TongAWuLLinQProteomic analysis of cellular protein alterations using a hepatitis B virus-producing cellular modelProteomics20088102012202318491315
- YangFYanSHeYExpression of hepatitis B virus proteins in transgenic mice alters lipid metabolism and induces oxidative stress in the liverJ Hepatol2008481121918037187
- LiHZhuWZhangLThe metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatmentSci Rep20155842125672227
- NaTYShinYKRohKJLiver X receptor mediates hepatitis B virus X protein-induced lipogenesis in hepatitis B virus-associated hepatocellular carcinomaHepatology20094941122113119105208
- YueDZhangYWChengLLHepatitis B virus X protein (HBx)-induced abnormalities of nucleic acid metabolism revealed by 1H-NMR-based metabonomicsSci Rep201662443027075403
- DaiYCrosMPPontoizeauCElena-HermannBBonnGKHainautPDownregulation of transcription factor E4F1 in hepatocarcinoma cells: HBV-dependent effects on autophagy, proliferation and metabolismCarcinogenesis201435363565024163401
- XieQFanFWeiWMulti-omics analyses reveal metabolic alterations regulated by hepatitis B virus core protein in hepatocellular carcinoma cellsSci Rep201774108928112229
- MinHKSookoianSPirolaCJChengJMirshahiFSanyalAJMetabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in Huh-7 hepatoma cellsAm J Physiol Gastrointest Liver Physiol20143071G66G7624763554
- FangMDewaeleSZhaoYPSerum N-glycome biomarker for monitoring development of DENA-induced hepatocellular carcinoma in ratMol Cancer2010921520704698
- HongEJLevasseurMPDufourCRPerryMCGiguereVLoss of estrogen-related receptor α promotes hepatocarcinogenesis development via metabolic and inflammatory disturbancesProc Natl Acad Sci U S A201311044179751798024127579
- TanYYinPTangLMetabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: potential biomarkers effective for small hepatocellular carcinoma diagnosisMol Cell Proteomics2012112M111.010694
- ZengJHuangXZhouLMetabolomics identifies biomarker pattern for early diagnosis of hepatocellular carcinoma: from diethylnitrosamine treated rats to patientsSci Rep201551610126526930
- HuangXZengJZhouLHuCYinPLinXA new strategy for analyzing time-series data using dynamic networks: identifying prospective biomarkers of hepatocellular carcinomaSci Rep201663244827578360
- LiZFWangJHuangCGas chromatography/time-of-flight mass spectrometry-based metabonomics of hepatocarcinoma in rats with lung metastasis: elucidation of the metabolic characteristics of hepatocarcinoma at formation and metastasisRapid Commun Mass Spectrom201024182765277520814984
- WangJZhangSLiZ1H-NMR-based metabolomics of tumor tissue for the metabolic characterization of rat hepatocellular carcinoma formation and metastasisTumour Biol201132122323120890798
- TengCFHsiehWCYangCWA biphasic response pattern of lipid metabolomics in the stage progression of hepatitis B virus X tumorigenesisMol Carcinog201655110511425594851
- FanTRongZDongJMetabolomic and transcriptomic profiling of hepatocellular carcinomas in Hras12V transgenic miceCancer Med20176102370238428941178
- LiSLiuHJinYLinSCaiZJiangYMetabolomics study of alcohol-induced liver injury and hepatocellular carcinoma xenografts in miceJ Chromatogr B Analyt Technol Biomed Life Sci20118792423692375
- AssiNFagesAVineisPA statistical framework to model the meeting-in-the-middle principle using metabolomic data: application to hepatocellular carcinoma in the EPIC studyMutagenesis201530674375326130468
- FagesADuarte-SallesTStepienMMetabolomic profiles of hepatocellular carcinoma in a European prospective cohortBMC Med20151324226399231
- ChenFXueJZhouLWuSChenZIdentification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic methodAnal Bioanal Chem201140161899190421833635
- ChenSKongHLuXPseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high-performance liquid chromatography/triple quadrupole mass spectrometryAnal Chem201385178326833323889541
- ZengZLiuXDaiWIon fusion of high-resolution LC-MS-based metabolomics data to discover more reliable biomarkersAnal Chem20148683793380024611595
- LiangQLiuHWangCLiBPhenotypic characterization analysis of human hepatocarcinoma by urine metabolomics approachSci Rep201661976326805550
- ZhangASunHYanGHanYYeYWangXUrinary metabolic profiling identifies a key role for glycocholic acid in human liver cancer by ultra-performance liquid-chromatography coupled with high-definition mass spectrometryClin Chim Acta2013418869023313056
- ChenTXieGWangXSerum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinomaMol Cell Proteomics2011107M110.004945
- NahonPAmathieuRTribaMNIdentification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosisClin Cancer Res201218246714672223136190
- RessomHWXiaoJFTuliLUtilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosisAnal Chim Acta20127439010022882828
- XiaoJFVargheseRSZhouBLC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohortJ Proteome Res201211125914592323078175
- LiuYHongZTanGNMR and LC-MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosisInt J Cancer2014135365866824382646
- ZengJYinPTanYMetabolomics study of hepatocellular carcinoma: discovery and validation of serum potential biomarkers by using capillary electrophoresis-mass spectrometryJ Proteome Res20141373420343124853826
- FitianAINelsonDRLiuCXuYAraratMCabreraRIntegrated metabolomic profiling of hepatocellular carcinoma in hepatitis C cirrhosis through GC-MS and UPLC-MS-MSLiver Int20143491428144424661807
- RanjbarMRLuoYDi PotoCGC-MS based plasma metabolomics for identification of candidate biomarkers for hepatocellular carcinoma in Egyptian cohortPLoS One2015106e012729926030804
- Di PotoCFerrariniAZhaoYMetabolomic characterization of hepatocellular carcinoma in patients with liver cirrhosis for biomarker discoveryCancer Epidemiol Biomarkers Prev201726567568327913395
- SanabriaJRKombuRSZhangGFGlutathione species and metabolomic prints in subjects with liver disease as biological markers for the detection of hepatocellular carcinomaHPB (Oxford)2016181297999028340971
- WangBChenDChenYMetabonomic profiles discriminate hepatocellular carcinoma from liver cirrhosis by ultraperformance liquid chromatography-mass spectrometryJ Proteome Res20121121217122722200553
- ShaoYZhuBZhengRDevelopment of urinary pseudotargeted LC-MS-based metabolomics method and its application in hepatocellular carcinoma biomarker discoveryJ Proteome Res201514290691625483141
- ShariffMIGomaaAICoxIJUrinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation studyJ Proteome Res20111041828183621275434
- OsmanDAliOObadaMEl-MezayenHEl-SaidHChromatographic determination of some biomarkers of liver cirrhosis and hepatocellular carcinoma in Egyptian patientsBiomed Chromatogr2017316e3893
- DaiWYinPChenPStudy of urinary steroid hormone disorders: difference between hepatocellular carcinoma in early stage and cirrhosisAnal Bioanal Chem2014406184325433524817358
- CaoHHuangHXuWFecal metabolome profiling of liver cirrhosis and hepatocellular carcinoma patients by ultra performance liquid chromatography-mass spectrometryAnal Chim Acta20116911–2687521458633
- PattersonADMaurhoferOBeyogluDAberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profilingCancer Res201171216590660021900402
- ZhouLDingLYinPSerum metabolic profiling study of hepatocellular carcinoma infected with hepatitis B or hepatitis C virus by using liquid chromatography-mass spectrometryJ Proteome Res201211115433544222946841
- TianSChangHHWangCJiangJWangXNiuJMulti-TGDR, a multi-class regularization method, identifies the metabolic profiles of hepatocellular carcinoma and cirrhosis infected with hepatitis B or hepatitis C virusBMC Bioinformatics2014159724707821
- BowersJHughesESkillNMaluccioMRafteryDDetection of hepatocellular carcinoma in hepatitis C patients: biomarker discovery by LC-MSJ Chromatogr B Analyt Technol Biomed Life Sci2014966154162
- WeiSSuryaniYGowdaGASkillNMaluccioMRafteryDDifferentiating hepatocellular carcinoma from hepatitis C using metabolite profilingMetabolites20122470171624957758
- BaniasadiHGowdaGAGuHTargeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MSElectrophoresis201334192910291723856972
- GaoRChengJFanCSerum metabolomics to identify the liver disease-specific biomarkers for the progression of hepatitis to hepatocellular carcinomaSci Rep201551817526658617
- GongZGZhaoWZhangJMetabolomics and eicosanoid analysis identified serum biomarkers for distinguishing hepatocellular carcinoma from hepatitis B virus-related cirrhosisOncotarget2017838638906390028969038
- LinXGaoJZhouLYinPXuGA modified k-TSP algorithm and its application in LC-MS-based metabolomics study of hepatocellular carcinoma and chronic liver diseasesJ Chromatogr B Analyt Technol Biomed Life Sci2014966100108
- ZhouLWangQYinPSerum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseasesAnal Bioanal Chem2012403120321322349331
- SogaTSugimotoMHonmaMSerum metabolomics reveals γ-glutamyl dipeptides as biomarkers for discrimination among different forms of liver diseaseJ Hepatol201155489690521334394
- HuangQTanYYinPMetabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomicsCancer Res201373164992500223824744
- LiuSYZhangRLKangHFanZJDuZHuman liver tissue metabolic profiling research on hepatitis B virus-related hepatocellular carcinomaWorld J Gastroenterol201319223423343223801834
- BudhuARoesslerSZhaoXIntegrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomesGastroenterology2013144510661075 e106123376425
- BeyoğluDImbeaudSMaurhoferOTissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classificationHepatology201358122923823463346
- MurakamiYKuboSTamoriAComprehensive analysis of transcriptome and metabolome analysis in intrahepatic cholangiocarcinoma and hepatocellular carcinomaSci Rep201551629426538415
- DarpolorMMBasuSSWorthAThe aspartate metabolism pathway is differentiable in human hepatocellular carcinoma: transcriptomics and 13C-isotope based metabolomicsNMR Biomed201427438138924497316
- SpratlinJLPittsTMKulikowskiGNSynergistic activity of histone deacetylase and proteasome inhibition against pancreatic and hepatocellular cancer cell linesAnticancer Res20113141093110321508352
- ZhengJFLuJWangXZGuoWHZhangJXComparative metabolomic profiling of hepatocellular carcinoma cells treated with sorafenib monotherapy vs. sorafenib-everolimus combination therapyMed Sci Monit2015211781179126092946
- AsgharUMeyerTAre there opportunities for chemotherapy in the treatment of hepatocellular cancer?J Hepatol201256368669521971559
- IwaoCShidojiYUpregulation of energy metabolism-related, p53-target TIGAR and SCO2 in HuH-7 cells with p53 mutation by geranylgeranoic acid treatmentBiomed Res201536637138126700591
- ChengMLShiaoMSChiuDTWengSFTangHYHoHYBiochemical disorders associated with antiproliferative effect of dehydroepian-drosterone in hepatoma cells as revealed by LC-based metabolomicsBiochem Pharmacol201182111549156121843511
- IkutaNChikamotoKAsanoYTime course effect of R-α-lipoic acid on cellular metabolomics in cultured hepatoma cellsJ Med Food201720321122228296595
- QinXYWeiFTanokuraMThe effect of acyclic retinoid on the metabolomic profiles of hepatocytes and hepatocellular carcinoma cellsPLoS One2013812e8286024376596
- NoguchiYYoungJDAlemanJOHansenMEKelleherJKStephanopoulosGTracking cellular metabolomics in lipoapoptosis- and steatosis-developing liver cellsMol Biosyst2011751409141921327189
- KumarPSinghAKRajV6,7-Dimethoxy-1,2,3,4-tetrahydro-isoquinoline-3-carboxylic acid attenuates hepatocellular carcinoma in rats with NMR-based metabolic perturbationsFuture Sci OA201733FSO20228884001
- LiFLiXMiaoYUHPLC-MS-based metabolomics analysis on mice bearing neoplasm (H22) for hispidulinJ Pharm Biomed Anal201612531031827077962
- MaTFanBYZhangCMetabonomics applied in exploring the antitumour mechanism of physapubenolide on hepatocellular carcinoma cells by targeting glycolysis through the Akt-p53 pathwaySci Rep201662992627416811
- QinXYTatsukawaHHitomiKMetabolome analyses uncovered a novel inhibitory effect of acyclic retinoid on aberrant lipogenesis in a mouse diethylnitrosamine-induced hepatic tumorigenesis modelCancer Prev Res (Phila)20169320521426744170
- BaoYWangSYangXLiTXiaYMengXMetabolomic study of the intervention effects of shuihonghuazi formula, a traditional Chinese medicinal formulae, on hepatocellular carcinoma (HCC) rats using performance HPLC/ESI-TOF-MSJ Ethnopharmacol201719846847828108381
- ZhouLLiaoYYinPMetabolic profiling study of early and late recurrence of hepatocellular carcinoma based on liquid chromatography-mass spectrometryJ Chromatogr B Analyt Technol Biomed Life Sci2014966163170
- YeGZhuBYaoZAnalysis of urinary metabolic signatures of early hepatocellular carcinoma recurrence after surgical removal using gas chromatography-mass spectrometryJ Proteome Res20121184361437222768978
- GoossensCNahonPle MoyecLSequential serum metabolomic profiling after radiofrequency ablation of hepatocellular carcinoma reveals different response patterns according to etiologyJ Proteome Res20161551446145427015127
- ChenYZhouJLiJFengJChenZWangXPlasma metabolomic analysis of human hepatocellular carcinoma: diagnostic and therapeutic studyOncotarget2016730473324734227322079
- SpratlinJLSerkovaNJEckhardtSGClinical applications of metabolomics in oncology: a reviewClin Cancer Res200915243144019147747
- GikaHGTheodoridisGAPlumbRSWilsonIDCurrent practice of liquid chromatography-mass spectrometry in metabolomics and metabonomicsJ Pharm Biomed Anal201487122523916607
- GrebeSKSinghRJLC-MS/MS in the clinical laboratory: where to from here?Clin Biochem Rev201132153121451775
- VermeerschKAStyczynskiMPApplications of metabolomics in cancer researchJ Carcinog201312923858297
- HwangVJWeissRHMetabolomic profiling for early cancer detection: current status and future prospectsExpert Opin Drug Metab Toxicol Epub2016913
- GoodacreRVaidyanathanSDunnWBHarriganGGKellDBMetabolomics by numbers: acquiring and understanding global metabolite dataTrends Biotechnol200422524525215109811
- NicholsonJKLindonJCSystems biology: metabonomicsNature200845572161054105618948945
- EverettJRPharmacometabonomics in humans: a new tool for personalized medicinePharmacogenomics201516773775425929853