Abstract
Purpose
The identification of prognostic markers for colorectal cancer (CRC) is needed for clinical practice. Fructose-bisphosphate aldolase A (ALDOA) and DEAD box p68 RNA helicase (DDX5) are commonly overexpressed in cancer and correlate with tumorigenesis. However, association between expression of ALDOA and DDX5, and CRC outcome has not been reported.
Patients and methods
We used 141 formalin-fixed paraffin-embedded (FFPE) specimens collected from 105 patients with CRC treated at the Affiliated Hospital of Guilin Medical University and the People’s Hospital of Liuzhou. We performed tissue microarray based immunohistochemistry to explore expression features and prognostic value (overall survival, OS; disease-free survival, [DFS]) of ALDOA and DDX5 in CRC tissues. The prognostic values were evaluated using Kaplan–Meier analysis, and Cox regression analyses.
Results
ALDOA and DDX5 were highly expressed in CRC tissues and liver metastatic CRC tissues compared with normal glandular epithelium tissues (all p<0.05). Interestingly, primary CRC tissues highly expressing ALDOA or DDX5 had poor outcome (p<0.0001 for both OS and DFS for ALDOA; p=0.001 for OS; and p=0.011 for DFS for DDX5) compared with patients who had low expression of those proteins. Furthermore, multivariate Cox analysis showed that ALDOA/DDX5 combination was an independent risk factor for OS and ALDOA was an independent risk factor for DFS.
Conclusion
High levels of ALDOA and DDX5 contribute to the aggressiveness and poor prognosis of CRC. ALDOA/DDX5 expression could be a biomarkers for the prognosis of CRC.
Introduction
Colorectal cancer (CRC) is a common malignancy of the digestive system and ranks fifth in China for both cancer incidence (376.3 per 100,000) and mortality (191.0 per 100,000).Citation1 Over the past decade, significant progress has been made in the treatment of CRC through advances in surgery, radiotherapy, chemotherapy, and targeted therapy.Citation2–Citation4 However, the majority of patients are diagnosed at an advanced stage, limiting the therapeutic options for improving the survival rate and leading to a poor prognosis.Citation5 In recent years, several prognostic markers such as SERPINA4,Citation6 c-MYC, and ß-cateninCitation7 have been proposed as immunohistochemical markers for CRC, while established diagnostic and prognostic serum biomarkers, including carbohydrate antigen 19-9 (CA19-9),Citation8 are of value in the treatment of CRC patients. However, the need remains to identify effective biomarkers that can classify patients at high or low risk of outcomes after surgical resection.
Fructose-bisphosphate aldolase A (ALDOA), an aldolase isozyme, plays a key role in glycolysis and gluconeogenesisCitation9 and is highly expressed in many types of cancers, including kidney, lung,Citation10 oral squamous cell,Citation11 and hepatocellular carcinomas.Citation12 DEAD box p68 RNA helicase (DDX5) is considered a prototypic member of the DEAD-box family of RNA helicases.Citation13 Recent studies have also demonstrated that DDX5 is aberrantly expressed in several types of cancers, including colon cancer,Citation14 breast cancer,Citation15 lung cancer,Citation16 cutaneous squamous cell carcinoma,Citation17 and suggesting that DDX5 plays important roles in cancer development and progression.Citation18 However, limited information was available for prognostic value of ALDOA and DDX5 in CRC.
In this study, we demonstrated that ALDOA is highly expressed in CRC and liver metastatic CRC tissues compared with paired normal glandular epithelium tissues, and could therefore represent a potential prognostic marker for primary CRC patients after surgical resection.
Materials and methods
Patient tissue samples
We evaluated 141 formalin-fixed paraffin-embedded (FFPE) specimens (105 CRC tissues, 18 paired normal glandular epithelium tissues, and 18 liver metastatic CRC tissues) collected from 105 patients with CRC treated at the Affiliated Hospital of Guilin Medical University and the People’s Hospital of Liuzhou from April 2012 to March 2015 with written informed consent by each patient and approval from the institutional review board of Affiliated Hospital of Guilin Medical University and the People’s Hospital of Liuzhou was obtained. H&E-stained slides were prepared from each FFPE specimen and were reviewed by experienced pathologists. The diagnosis of CRC was confirmed based on clinical manifestation, and pathological and serological examinations.
Follow-up
Overall survival (OS) was calculated from the date of surgery to the date of death or the last known follow-up. Disease-free survival (DFS) was calculated from the date of tumor resection until the detection of tumor recurrence, metastasis, or death. All patients enrolled in this study provided written informed consent and the protocol was approved by the ethics committees of the Affiliated Hospital of Guilin Medical University and the People’s Hospital of Liuzhou. No patients had received radiotherapy, chemotherapy, hormone therapy, or other related anti-tumor therapies prior to surgery. Follow-up data were summarized at the end of October 2016. Follow-up of all of the patients was carried out at both hospitals, including tumor marker testing every 3 months and diagnostic imaging at least every 6 months, based on the surveillance suggested in the guidelines. In cases of suspected recurrence, MRI or CT were included in the diagnostic imaging.Citation19
Tissue microarrays and immunohistochemistry
Tissue microarrays were constructed from a representative core from each FFPE specimen. Tissue cylinders with a diameter of 1.5 mm were punched from marked areas of each sample and incorporated into a recipient paraffin block. Sections of 4 μm thickness were placed on slides coated with 3-aminopropyltriethoxysilane. All H&E-stained slides were reviewed by experienced pathologists and the representative cores were pre-marked in the paraffin blocks.Citation20
Paraffin sections were deparaffinized in xylene and rehydrated using decreasing concentrations of ethanol (100%, 95%, and 85% for 5 min each). Antigen retrieval was performed by microwave irradiation for 5 min in pH 6.0 citric buffer, after which the samples were cooled at room temperature for 60 min. Endogenous peroxidase activity was blocked by incubation of the slides in 3% H2O2/phosphate-buffered saline, and non-specific binding sites were blocked with goat serum.Citation20 Mouse monoclonal antibody to ALDOA (H00000226-M02, Abnova, Taipei City, Taiwan; 1:200 dilution) and rabbit monoclonal antibody to DDX5 (ab126730, Abcam, Cambridge, UK; 1:400 dilution) were used as a primary antibody, and an EnVision Detection Kit (GK500705: Gene Tech, Shanghai, People’s Republic of China) was used to visualize tissue antigens. Tissue sections were counterstained with hematoxylin for 5 min. Negative control slides without primary antibody were prepared for all assays. Imaging was performed using a Leica CCD DFC420 camera connected to a Leica DM IRE2 microscope (Leica Microsystems Imaging Solutions Ltd, Cambridge, UK). Photographs of representative fields were captured under high-power magnification (×200) using Leica QWin Plus v3 software. The integrated optical density (IOD) of each image was measured using Image-Pro Plus v6.0 software (Media Cybernetics Inc, Bethesda, MD, USA).Citation20 Expression levels of ALDOA and DDX5 were thus represented by IOD.Citation20
Statistical analysis
X2 analyses, correlations between variables, Kaplan–Meier analyses, univariate survival analysis, and multiple Cox proportional hazards regression were performed using the SPSS statistical software package (SPSS Standard version 13.0; SPSS, Chicago, IL, USA). The optimum cut-off points for ALDOA and DDX5 expression were obtained using X-tile software version 3.6.1 (Yale University School of Medicine, New Haven, CT, USA) to determine the relationship between protein expression and clinical outcomes (OS and DFS).Citation21 A significant difference was considered if the p-value from a two-tailed test was <0.05.
Results
Expression levels of ALDOA and DDX5 in paired adjacent glandular epithelium, CRC tumor tissues, and liver metastatic CRC tissues
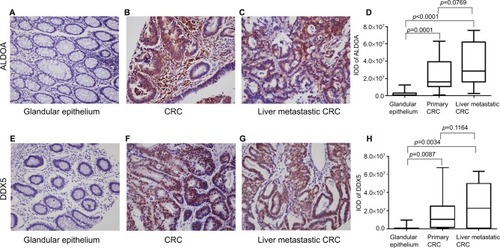
ALDOA expression in adjacent glandular epithelium was negative or low (), but high expression was observed in CRC tumor tissue () and liver metastatic CRC tissue (). Significant statistical differences were observed between the paired adjacent normal glandular epithelium tissue and the CRC tumor tissue (n=18, p=0.0001, ), and also between the paired adjacent normal glandular epithelium tissue and the liver metastatic CRC tissue (n=18, p<0.0001, ). In addition, ALDOA was highly expressed in liver metastatic CRC tissue compared with paired primary CRC tissue (p=0.0769). In a parallel manner, DDX5 expression was significantly higher in CRC tumor tissue (; n=18, p=0.0034, ) and liver metastatic CRC tissue (; n=18, p=0.0087, ) compared with tissues of adjacent glandular epithelium ().
Figure 1 Expression of ALDOA and DDX5 in paired adjacent glandular epithelium, primary colorectal cancer, and liver metastatic colorectal cancer tissue.
Notes: Expression of ALDOA in (A) adjacent glandular epithelium (×200), (B) primary colorectal cancer tissue (×200), (C) liver metastatic colorectal cancer tissue (×200). (D) Box plot showing the staining intensity (mean with SEM) of ALDOA in paired adjacent glandular epithelium, primary colorectal cancer, and liver metastatic colorectal cancer tissues. Expression of DDX5 in (E) adjacent glandular epithelium (×200), (F) primary colorectal cancer tissue (×200), (G) liver metastatic colorectal cancer tissue (×200). (H) Box plot showing the staining intensity (mean with SEM) of ALDOA in paired adjacent glandular epithelium, primary colorectal cancer, and liver metastatic colorectal cancer tissues.
Abbreviations: ALDOA, fructose-bisphosphate aldolase A; CRC, colorectal cancer; DDX5, DEAD box p68 RNA helicase; IOD, integrated optical density.
High expression of ALDOA and DDX5 are associated with poor prognosis
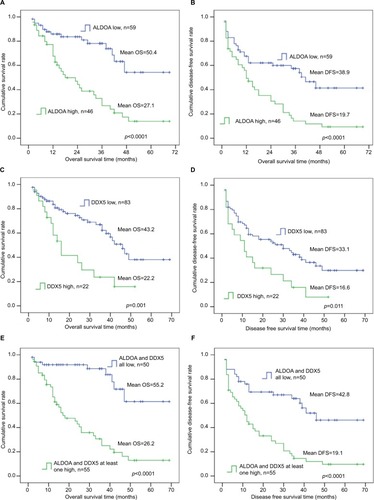
According to the optimum cut-off point obtained from X-tile analyses, OS and DFS were estimated by Kaplan–Meier analyses. Forty-six patients were classified in the high ALDOA expression group and 59 were classified in the low ALDOA expression group; 83 patients were classified in the low DDX5 expression group and 22 in the high expression group. Kaplan–Meier analyses clearly showed that CRC patients with higher expression of ALDOA had worse OS and DFS compared with patients with lower expression of ALDOA (p<0.0001 for both OS and DFS; ); higher expression of DDX5 had worse OS and DFS, compared with patients with lower expression of DDX5 (p=0.001 for OS and p=0.011 for DFS; ). Notably, when combining ALDOA and DDX5, the prognostic values were more significant for predicting OS and DFS ().
Figure 2 Positive correlation of high ALDOA expression and DDX5 high expression with poor OS and DFS in colorectal cancer patients.
Notes: Probabilities of (A) OS (high ALDOA=46, low ALDOA=59, p<0.0001) and (B) DFS (p<0.0001) in colorectal cancer patients. Probabilities of (C) OS (high DDX5=22, low DDX5=83, p=0.001) and (D) DFS (p=0.011) in colorectal cancer patients. Probabilities of (E) OS (ALDOA and DDX5 all low, n=50, ALDOA and DDX5 at least one high, n=55, p<0.0001) and (F) DFS (p<0.0001) in colorectal cancer patients. Log-rank test was determined using Kaplan–Meier survival analyses.
Abbreviations: ALDOA, fructose-bisphosphate aldolase A; DDX5, DEAD box p68 RNA helicase; DFS, disease-free survival; OS, overall survival.
Association of ALDOA and DDX5 expression with clinicopathological features of CRC patients
We further investigated the association between ALDOA and DDX5 expression and clinicopathological factors in the 105 CRC patients. X2 analyses showed that DDX5 was not associated with any clinicopathological factors, while ALDOA expression was associated with serum CA19-9 level in CRC patients (p=0.027) but was not associated with sex, age, tumor differentiation, T stage, N stage, M stage, TNM stage, serum carcinoembryonic antigen (CEA), liver metastasis, or postoperative chemotherapy ().
Table 1 Relationship between ALDOA and DDX5 expression levels, and the clinicopathological features of CRC
Univariate and multivariate Cox regression analysis of risk factors for CRC patients after surgery
To determine independent prognostic factors for CRC patients after surgery, univariate and multivariate analyses were performed based on a Cox proportional hazard regression model. Clinical and pathological factors showing statistical significance in Cox univariate analyses were included in Cox multivariate analyses. Cox univariate analyses showed that M stage (p<0.0001 for both OS and DFS), TNM stage (p=0.005 for OS and p<0.0001 for DFS), serum CA19-9 (p<0.0001 for OS, p=0.001 for DFS), liver metastasis (p<0.0001 for both OS and DFS), ALDOA expression (p<0.0001 for OS, p=0.001 for DFS), DDX5 expression (p=0.002 for OS, p=0.015 for DFS), and ALDOA/DDX5 combination (p<0.0001 for both OS and DFS) were prognostic factors, and serum CA19-9 (p=0.004 for OS, p=0.005 for DFS), liver metastasis (p<0.0001 for both OS and DFS), and ALDOA/DDX5 combination (p<0.0001 for OS) and ALDOA expression (p=0.001 for DFS) were independent prognostic factors in CRC patients after surgery ().
Table 2 Univariate and multivariate analyses of factors associated with OS and DFS in CRC patients
Discussion
CRC is the most commonly diagnosed cancer and a major cause of global cancer death in both males and females.Citation22 The TNM staging system is a conventional predictor of outcome among postoperative CRC patients. Despite continuous refinement of the system to indicate the extent of the disease and define prognosis, with the aim of guiding treatment, OS and DFS among postoperative CRC patients may vary considerably, even within the same tumor stage.Citation23 Therefore, the need for novel biomarkers, especially those which might reflect tumor features to more precisely stratify patients into different risk categories, is clearly warranted.Citation24
Although serum CEA and CA19-9 are elevated in patients with CRC, they have not have not been recommended as screening tests for CRC, due to their low sensitivity and specificity. Also, they were found to be significantly elevated in colorectal neoplasia.Citation25–Citation27 However, serum CEA and CA19-9 are recommended as a prognostic biomarker for monitoring recurrence of CRC following curative resection and predicting prognosis.Citation25,Citation28 Univariate analysis of the present study reveals that serum CEA had a tendency to predict prognosis and serum CA19-9 is an independent prognostic factor for both OS and DFS.
Immunohistochemical staining is widely used for visualizing these tumor markers and is a routine tool in the department of pathology. Although several studies have reported diagnostic and prognostic markers of CRC for immunohistochemistry assays, few are routinely used in clinical pathology. Thus, identification of a new molecular marker for use in immunohistochemistry of tumor tissues from CRC patients would be beneficial for effective individualized therapy for this disease.
In this present study, we initially evaluated the significance of ALDOA and DDX5 expressions in 18 paired normal glandular epithelium tissues, primary CRC tissues, and liver metastatic CRC tissues. The paired t-test revealed that ALDOA and DDX5 expression were significantly higher in primary CRC tissues and liver metastatic CRC tissues than in normal glandular epithelium tissues. Furthermore, ALDOA and DDX5 expression were slightly higher in liver metastatic CRC tissues than in primary tissues, although the small sample size meant that the difference was not statistically significant (p=0.0769 for ALDOA and p=0.1164 for DDX5). Further studies in a larger sample of paired liver metastatic CRC and primary tissues are therefore warranted.
More importantly, Kaplan–Meier survival analyses clearly showed that patients with high ALDOA or high DDX5 in expression in primary CRC tissues had shorter OS and DFS than patients with low ALDOA or low DDX5 expressions. Univariate and multivariate Cox regression analyses showed that serum CA19-9, liver metastasis, and ALDOA/DDX5 combination was an independent prognostic factor for OS, and serum CA19-9, liver metastasis, ALDOA were independent prognostic factors for DFS. Notably, multivariate Cox regression analyses showed that hazard ratio and p-value from multivariate Cox regression analyses reveals that ALDOA/DDX5 combination was more effective for predicting postoperative OS of CCR patients. Of note, dynamic proteomic analysis has recently identified ALDOA as a novel biomarker of CRC prognosisCitation29 and DDX5 was reported as a transcriptional co-activator in tumor development;Citation30,Citation31 however, detailed information on the prognostic value of ALDOA and DDX5 and its expression characteristics in liver metastatic CRC tissues has not been reported.
To the best of our knowledge, our study is the first to demonstrate the prognostic value of ALDOA, DDX5, and ALDOA/DDX5 combination as an independent prognostic factor for OS and DFS in CRC patients after surgery. Furthermore, ALDOA and or ALDOA/DDX 5 combined with serum CA19-9, liver metastasis may effectively predict outcome of CRC patients.
Conclusion
The present study clearly demonstrates that ALDOA and DDX5 protein are highly expressed in CRC tumor and liver metastatic CRC tissues compared with normal glandular epithelium tissues, and that increased expression of ALDOA and DDX5 in CRC tissues may indicate poor prognosis in patients after surgery. Therefore, our findings indicate that ALDOA and DDX5 are potential biomarkers for CRC, and may represent a useful approach to the prediction of OS and DFS in CRC patients after surgery.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81360326, 81260354, 81460458), Guangxi Natural Science Foundation of China (2014GXNS-FAA118219, 2015GXNSFAA139157), Bagui Scholar Foundation of He Songqing, and the “Sphingolipids and Related Diseases” Programme for the Innovative Research Team of Guilin Medical University. We thank Clare Cox from Liwen Bianji, Edanz Group China for editing the English text of a draft of this manuscript.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- ChenWZhengRBaadePDCancer statistics in China, 2015CA Cancer J Clin201666211513226808342
- GibsonTBRanganathanAGrotheyARandomized phase III trial results of panitumumab, a fully human anti-epidermal growth factor receptor monoclonal antibody, in metastatic colorectal cancerClin Colorectal Cancer200661293116796788
- HurwitzHIFehrenbacherLHainsworthJDBevacizumab in combination with fluorouracil and leucovorin: an active regimen for first-line metastatic colorectal cancerJ Clin Oncol200523153502350815908660
- WolpinBMMeyerhardtJAMamonHJMayerRJAdjuvant treatment of colorectal cancerCA Cancer J Clin200757316818517507442
- TokunagaRSakamotoYNakagawaSPrognostic nutritional index predicts severe complications, recurrence, and poor prognosis in patients with colorectal cancer undergoing primary tumor resectionDis Colon Rectum201558111048105726445177
- SunHMMiYSYuFDSERPINA4 is a novel independent prognostic indicator and a potential therapeutic target for colorectal cancerAm J Cancer Res2016681636164927648355
- LeeKSKwakYNamKHFavorable prognosis in colorectal cancer patients with co-expression of c-MYC and ss-cateninBMC Cancer201616173027619912
- FilellaXMolinaRGrauJJPrognostic value of CA 19.9 levels in colorectal cancerAnn Surg1992216155591632702
- EspositoGVitaglianoLCostanzoPHuman aldolase A natural mutants: relationship between flexibility of the C-terminal region and enzyme functionBiochem J.2004380Pt 1515614766013
- KukitaAYoshidaMCFukushigeSMolecular gene mapping of human aldolase A (ALDOA) gene to chromosome 16Hum Genet198776120263570299
- OparinaNYSnezhkinaAVSadritdinovaAFDifferential expression of genes that encode glycolysis enzymes in kidney and lung cancer in humansGenetika201349781482324450150
- LessaRCCamposAHFreitasCEIdentification of upregulated genes in oral squamous cell carcinomasHead Neck201335101475148122987617
- ShimizuTInoueKHachiyaHShibuyaNShimodaMKubotaKFrequent alteration of the protein synthesis of enzymes for glucose metabolism in hepatocellular carcinomasJ Gastroenterol20144991324133224203292
- DaiTYCaoLYangZCP68 RNA helicase as a molecular target for cancer therapyJ Exp Clin Cancer Res2014336425150365
- SinghCHainesGKTalamontiMSRadosevichJAExpression of p68 in human colon cancerTumour Biol19951652812897652466
- HainesGKCajulisRHaydenRDudaRTalamontiMRadosevichJAExpression of the double-stranded RNA-dependent protein kinase (p68) in human breast tissuesTumour Biol19961715127501973
- Dosaka-AkitaHHaradaMMiyamotoHKawakamiYClinical significance of oncogene product expression in human lung cancerNihon Kyobu Shikkan Gakkai zasshi1992308144114471331597
- WangSJZhangCYouYShiCMOverexpression of RNA helicase p68 protein in cutaneous squamous cell carcinomaClin Exp Dermatol201237888288822548649
- Fuller-PaceFVMooreHCRNA helicases p68 and p72: multifunctional proteins with important implications for cancer developmentFuture Oncol20117223925121345143
- TanNLiuQLiuXLow expression of B-cell-associated protein 31 in human primary hepatocellular carcinoma correlates with poor prognosisHistopathology201668222122925980696
- CampRLDolled-FilhartMRimmDLX-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimizationClin Cancer Res200410217252725915534099
- ParkinDMBrayFFerlayJPisaniPGlobal cancer statistics, 2002CA Cancer J Clin20055527410815761078
- MaoYPXieFYLiuLZRe-evaluation of 6th edition of AJCC staging system for nasopharyngeal carcinoma and proposed improvement based on magnetic resonance imagingInt J Radiat Oncol Biol Phys20097351326133419153016
- PuppaGSonzogniAColombariRPelosiGTNM staging system of colorectal carcinoma: a critical appraisal of challenging issuesArch Pathol Lab Med2010134683785220524862
- LockerGYHamiltonSHarrisJASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancerJ Clin Oncol200624335313532717060676
- BagariaBSoodSSharmaRLalwaniSComparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis)Cancer Biol Med201310314815724379990
- KimNHLeeMYParkJHSerum CEA and CA 19-9 levels are associated with the presence and severity of colorectal neoplasiaYonsei Med J201758591892428792134
- YuZChenZWuJLiZWuYPrognostic value of pretreatment serum carbohydrate antigen 19-9 level in patients with colorectal cancer: a meta-analysis20171211e0188139
- PengYLiXWuMNew prognosis biomarkers identified by dynamic proteomic analysis of colorectal cancerMol Biosyst20128113077308822996014
- NicolSMFuller-PaceFVAnalysis of the RNA helicase p68 (Ddx5) as a transcriptional regulatorMethods Mol Biol201058726527920225156
- SambasivanRCheedipudiSPasupuletiNSalehAPavlathGKDhawanJThe small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoterJ Cell Sci2009122Pt 193481349119723804
