Abstract
Background and aim
Several dysregulated microRNAs (miRNAs) have been implicated in the pathogenesis of cholangiocarcinoma (CCA); however, small sample sizes and invariable research designs are limitations, hindering a thorough analysis of miRNAs as diagnostic and prognostic tools for CCA. This study aimed to systematically summarize the clinical value of miRNAs in human CCA both for all available miRNAs and single miRNA with multiple researches.
Methods
Pooled parameters included the area under the curve (AUC), sensitivity, specificity, and hazard ratios (HRs) to separately determine overall diagnostic and prognostic performance. Subgroup and sensitivity analyses were performed only in the event of heterogeneity. Thirty-four studies including 12 diagnostic studies and 22 prognostic studies were eligible for inclusion in this meta-analysis.
Results
We observed that miR-21, miR-26, miR-483, miR-106a, miR-150, miR-192, and miR-194 were employed for distinguishing patients with CCA from healthy controls. Pooled sensitivity, specificity, and AUC were 0.82 (95% confidence interval [CI] 0.77–0.86), 0.83 (95% CI 0.75–0.89), and 0.88 (95% CI 0.85–0.91), respectively. Abnormal expression of miR-21, miR-26a, miR-192, miR-200c, miR-221, miR-29a, miR-191, miR-181c, miR-34a, miR-106a, miR-203, and miR-373 in patients was confirmed to associate with poor survival rate. Pooled HRs and 95% CIs were calculated using STATA, resulting in the pooled HR of 1.47 (95% CI 0.91–2.37) for overall survival (OS), 0.67 (95% CI 0.16–2.81) for disease-free survival (DFS), 2.31 (95% CI 1.59–3.36) for progression-free survival (PFS), and 2.68 (95% CI 0.88–8.15) for relapse-free survival (RFS). Thus, CCA patients with dysregulated miRNA expression were confirmed to have shorter OS, DFS, PFS, and RFS. Data regarding the diagnostic and prognostic roles of miR-21 suggested pooled diagnostic results of miR-21 for sensitivity, specificity, and AUC were 0.85 (95% CI 0.76–0.91), 0.92 (95% CI 0.81–0.97), and 0.93 (95% CI 0.91–0.95), respectively, suggesting better diagnostic performance of miR-21 compared with other miRNAs. Meanwhile, pooled prognostic result of miR-21 for HR was 1.88 (95% CI 1.41–2.51), indicating miR-21 could more appropriately predict shorter OS in patients with CCA.
Conclusion
miRNAs may provide a new approach for clinical application, and miR-21 may be a promising biomarker for diagnosis and prognosis of CCA.
Introduction
Cholangiocarcinoma (CCA) is a rare biliary malignancy prone to lymphatic metastasis. CCA is increasingly common, and currently, it is the most frequent primary hepatic malignancy.Citation1,Citation2 In some Asian countries, the elevated incidence of CCA appears to correlate with liver fluke Opisthorchis viverrini (Ov) infections. Liver fluke infections subsequently result in chronic inflammation of the biliary tree and malignant transformation.Citation3,Citation4 Although the accuracy of current diagnostic methods for cancer has greatly improved, most patients with CCA are diagnosed at an unresectable stage of the disease, leading to a 5-year overall survival (OS) of <30%.Citation5 There are currently only few diagnostic techniques for detecting early-stage CCA, and also, biomarker studies available are limited;Citation6,Citation7 as such, novel biomarkers for diagnosis and prognosis are needed.Citation8
MicroRNAs (miRNAs) are endogenous small (~22 nt) noncoding RNAs that contribute to cell fate determination, proliferation, and cell death.Citation9–Citation11 Several studies indicate that miRNAs can be stably expressed in human plasma and serum, and miRNAs have also been demonstrated to be abnormally expressed in the circulatory system during tumorigenesis and inflammation.Citation12,Citation13 Kishimoto et alCitation14 found that miR-21 was overexpressed in bile duct cancer (accuracy, 0.89; sensitivity, 84%; and specificity, 98%), indicating that it may be used as a biomarker for distinguishing bile duct cancer patients from normal subjects. Silakit et alCitation15 confirmed high expression of miR-192 in sera of CCA patients, which predicted worse OS compared with those with low miR-192 expression. CA19-9 has been frequently used to diagnose and predict CCA patient outcomes.Citation16 However, low sensitivity or low specificity prevents CA19-9 from being adopted globally.Citation17 Therefore, identifying miRNAs unique to CCA may provide suitable biomarkers for detection and prognosis.
Methods
Search strategy
We searched PubMed, EMBASE, and the Web of Science for studies of associations between miRNAs and CCA patients. We used these search terms: (microRNA OR miRNA OR miR) and (cholangiocarcinoma OR bile duct cancer) and (diagnosis OR prognosis). We screened titles, abstracts, author information, and results. Studies were included if they focused on miRNAs in CCA patients identified by diagnosis of histopathological confirmation, provided information about the relationship between miRNAs and diagnosis (summary receiver operating characteristic [SROC] curves or sensitivity/specificity) or prognosis (hazard ratio [HR] with 95% confidence interval [CI] or Kaplan–Meier curves), included healthy individuals as controls, and had clearly defined miRNA thresholds. Studies were excluded if they met the following criteria: reviews, letters, commentaries, or meeting records; duplicate publications; used cell lines; combined two or more miRNAs to calculate overall sensitivity/specificity or HR; and lacked sufficient data to evaluate HR and 95% CIs. Three individual investigators screened and evaluated retrieved articles. Disagreement was resolved with detailed discussion.
Quality assessment
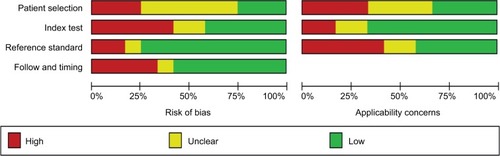
This meta-analysis had diagnostic and prognostic components, so the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) and the Meta-analysis of Observational Studies in Epidemiology group (MOOSE) were used to evaluate diagnostic and prognostic accuracy of eligible studies.Citation18,Citation19 If studies did not meet these criteria, they were excluded. Bias risk and applicability concerns are detailed in .
Data extraction
Two investigators independently examined each eligible study and its data using a standard protocol. We assessed first author, year of publication, study design, subject number, tumor stage, method of quantifying miRNA expression, and miRNA thresholds to stratify high- and low-subject groups. For diagnostic studies, sensitivity, specificity, true positives (TPs), false positives (FPs), false negatives (FN), and true negatives (TN) were included. For prognostic studies, HRs for OS or disease-free survival (DFS) or progression-free survival (PFS) or relapse-free survival (RFS), and 95% CIs and p values were extracted. If HR and relevant parameters were not available, we estimated HRs and 95% CIs using the methods of Parmar et al.Citation20
Statistical analysis
For diagnostic analysis, TPs, FPs, FNs, and TNs were extracted from included studies or recalculated based on the prevalence and sample size of each study. A bivariate model was used to calculate pooled sensitivity, specificity, positive and negative likelihood ratios (LRs), diagnostic odds ratios (DORs), and their 95% CIs.Citation21 SROC curves were plotted by pooling sensitivity and specificity of each study to evaluate diagnostic effects, along with auto-generation of AUCs of SROC curves and the maximum point of intersection between sensitivity and specificity (Q value). Heterogeneity was examined using an I2 test (p < 0.1 or I2 > 50% indicated significant heterogeneity).Citation22 Subgroup analysis was performed to identify potential heterogeneity sources, and Deeks’ funnel plot was used to inspect publication bias (p < 0.1 indicated statistically significant publication bias).
For prognostic analysis, multivariate analysis was employed to avoid confounding of exposure effects.Citation23 Heterogeneity assessment of combined HRs was performed using Cochran’s Q test and Higgins I2 statistic (p < 0.10 and/or I2 > 50% meant significant heterogeneity and the need for a fixed model). Otherwise, a random-effects model was used, and subgroup analysis was conducted to identify the source of heterogeneity. An observed HR > 1 indicated elevations of miRNA were detrimental and correlated with worse survival, but HR < 1 suggested lows of miRNA were harmful and related to poor survival. Furthermore, Begg’s funnel plots and Egger’s linear regression tests were used to evaluate publication bias.Citation24 All analyses was performed with Stata 12.0 (Stata Corporation, College Station, TX, USA).
Results
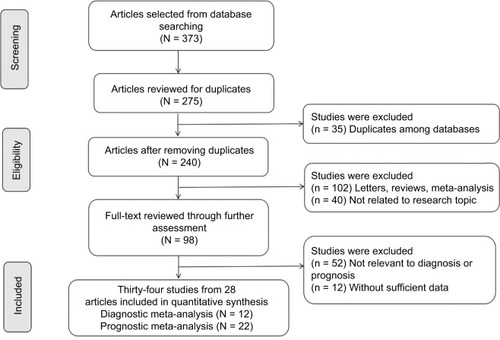
As shown in , a comprehensive search was performed on 30/04/2017, yielding a total of 373 results from the PubMed, EMBASE, and Web of Science. Among these results, 275 articles were identified as duplicates and irrelevant studies including reviews, textbooks, letters, or animal research after the screening of titles and abstract. The remaining 98 articles meeting the inclusion criteria were further thoroughly assessed. Finally, we found that 64 articles lacked sufficient data to allow for a meta-analysis or were not relevant to diagnoses or prognoses. Therefore, 28 articles containing 34 studies (including 12 for diagnostic analysis and 22 for prognostic analysis) focusing on the relationship between miRNAs and CCA were utilized for the final analysis. The main characteristics of each article are summarized in and .
Table 1 Main characteristics of the diagnostic studies included in the meta-analysis
Table 2 Main characteristics of the prognostic studies included in the meta-analysis
Figure 2 A flowchart of literature search and study selection. Twenty-three case–control studies including 9 studies for diagnoses and 14 studies for prognoses were included in this meta-analysis.
Diagnostic and prognostic value of miRNAs for CCA
Twelve studies containing 552 patients and 380 healthy controls evaluated the diagnostic value of miRNAs for CCA. Various miRNAs were detected and shown to be associated with the diagnosis of CCA, including miR-21 (n = 5), miR-192 (n = 2), miR-26 (n = 1), miR-194 (n = 1), miR-106a (n = 1), miR-150 (n = 1), and miR-482 (n = 1). Out of all the investigations, eight studies were conducted in Asian populations, and the remaining four studies were performed in Caucasian populations. Different samples including serum (n = 6), plasma (n = 3), urine (n = 2), and tissue (n = 1) were used in all these different studies. All studies used quantitative reverse transcription PCR (qRT-PCR) to measure the expression of miRNAs. Sensitivity, specificity, and DOR data are shown in , indicating that miRNAs have relatively high diagnostic value for CCA. I2 data are shown in . Meta-regression analysis revealed that plasma may be a source of heterogeneity, while other covariates, such as RNA extraction, measurements, specimen, and different races, may not contribute to heterogeneity (all p > 0.05) (). Pooled positive and negative LR data are shown in . Studies that included 1,155 patients from the People’s Republic of China, Italy, and Thailand were collected, and 12 miRNAs were identified in CCA patients. Tissue and sera were sample sources, and 13 reports assessed the correlations between abnormal miRNA expression and OS, while four studies assessed the relationship between miRNAs expression and DFS. Limited studies were included for PFS (n = 3) and RFS (n = 2); thus, pooled HRs were respectively analyzed in the systemic review but without further heterogeneity analysis.
Table 3 Multivariate meta-regression analysis for the associations of miRNAs with susceptibility to various cancers
Figure 3 SROC curves describing the diagnostic performance of miRNAs in discriminating cancer patients from healthy subjects: (A) miRNAs and (B) miR-21.
Abbreviations: AUC, area under the curve; miRNA, microRNA; SENS, sensitivity; SPEC, specificity; SROC, summary receiver operating characteristic.
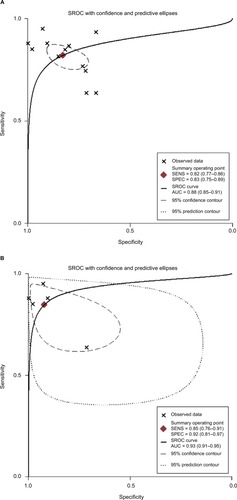
Figure 4 Forest plots of sensitivity and specificity of each included publication: (A) miRNAs and (B) miR-21.
Abbreviations: CI, confidence interval; miRNA, microRNA.
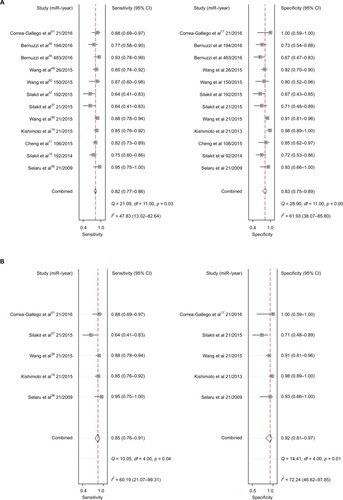
Figure 5 Graph of Deeks’ funnel plot asymmetry test: (A) miRNAs (p = 0.30) and (B) miR-21 (p = 0.64).
Abbreviations: ESS, effective sample size; miRNA, microRNA.
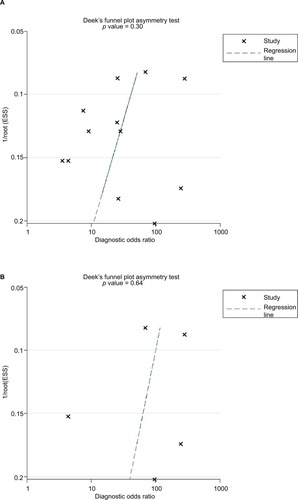
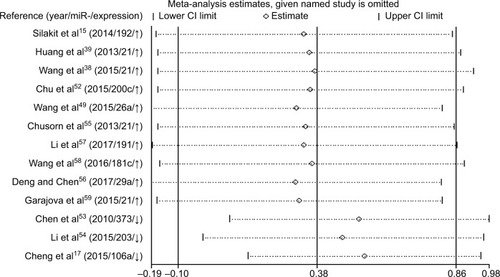
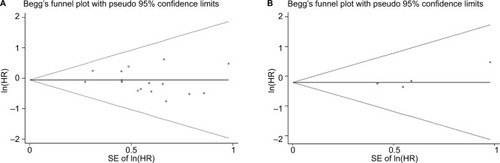
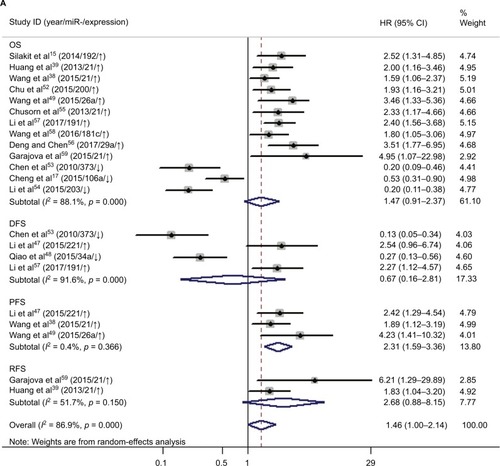
Increased expression of miR-21, miR-26a, miR-192, miR-200c, miR-191, miR-181c, miR-29a, and miR-221 was associated with a poor prognosis, as was decreased expression of miR-34a, miR-106a, miR-203, and miR-373 (). Heterogeneity data are shown in . A random-effects model was applied for the OS, DFS, and RFS subgroups, and the data are given in . Through subgroup analysis of OS determined by different elements, we found that the up–downregulation of miRNAs might be the source of heterogeneity (p = 0.001), while other factors (such as sample type, sample size, RNA extraction, and different measurements) may not lead to significant heterogeneity (all p > 0.05) (). The pooled results indicated that miRNAs were significant prognostic biomarkers for CCA patients (OS: HR = 1.47, 95% CI 0.91–2.37, n = 13; DFS: HR = 0.67, 95% CI 0.16–2.81, n = 4; PFS: HR = 2.31, 95% CI 1.59–3.36, n = 3; RFS: HR = 1.46, 95% CI 1.00–2.14, n = 2). Sensitivity analysis data are shown in . No obvious publication bias was found in the quantitative synthesis when Begg’s funnel plot and Egger’s test were applied to assess publication bias (p = 0.326) ().
Table 4 Subgroup analysis for association of miRNAs with OS in CCA
Figure 6 Forest plots of studies evaluating miRNAs expression level and cancer prognosis: (A) miRNAs and (B) miR-21.
Note: “↑” and ”↓” indicate that elevated and decreased expression of microRNAs correlate with poor survival rate.
Abbreviations: CI, confidence interval; DFS, disease-free survival; HR, hazard ratio; miRNA, microRNA; OS, overall survival; PFS, progression-free survival; RFS, relapse-free survival.
Diagnostic and prognostic values of miR-21 for CCA
Notably, we found that abnormal expression of miR-21 was identified by five studies for diagnostic analysis and four studies for prognostic analysis of all included studies. Thus, we had to conduct a meta-analysis for evaluating the potential diagnostic and prognostic value of miR-21 for CCA.
Five studies containing 235 patients assessed the diagnostic value of miR-21 for CCA. The pooled sensitivity, specificity, positive LR (PLR), negative LR (NLR), and DOR were 0.85 (95% CI 0.76–0.91), 0.92 (95% CI 0.81–0.97), 10.9 (95% CI 4.0–29.5), 0.17 (95% CI 0.10–0.28), and 66 (95% CI 16–272), respectively, indicating that miR-21 has a higher diagnostic accuracy than the pooled miRNAs for CCA patients (). Additionally, the AUC was 0.93 (95% CI 0.91–0.95), indicating miR-21 has a stronger diagnostic value for CCA when compared with the pooled miRNAs (). Furthermore, there was no significant publication bias in this meta-analysis ().
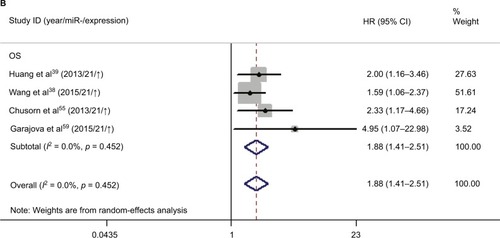
Meanwhile, we performed the prognostic analysis on the relationship of miR-21 and the OS of CCA patients (n = 4) (). As heterogeneity among all studies was not observed (OS: I2 = 0.0%; p = 0.452), a fixed-effects model was used. We found significant association between abnormal miR-21 expression and poor OS (HR = 1.88, 95% CI 1.41–2.51). Publication bias was evaluated with funnel plots and Begg’s tests. No significant publication biases were found ().
Discussion
CCA is the second most common primary hepatic malignancy arising from the bile duct epithelium,Citation7,Citation25 but it has few distinguishable symptoms, and patients are mostly diagnosed at the advanced tumor phase, resulting in the stubbornly high mortality in the recent years.Citation26–Citation28 Although the OS of patients with CCA is dismal, there is a large discrepancy between patients diagnosed at early and late stages, indicating an urgent need for novel diagnostic and prognostic biomarkers.Citation29
Current biomarkers for CCA have low sensitivity and specificity. Ince et alCitation30 constructed a receiver operating characteristic (ROC) curve and collected optimal thresholds for serum and biliary CA19-9 (serum: sensitivity, 49%; specificity, 84.5%; accuracy, 64%; biliary: sensitivity, 74%; specificity, 34%; accuracy, 69%) and noted the low sensitivity and specificity of these markers. They also reported sensitivity and specificity of biliary carcinoembryonic antigen (CEA) of 57% and 68%, respectively. Thus, CEA may be not ideal for distinguishing CCA from normal people. Huang et alCitation31 identified elevated soluble fragments of cytokeratin 19 (CYFRA 21-1) from CCA patients sera and reported an AUC of 0.879 and a high specificity (96.2%) but a low sensitivity (75.6%). Therefore, traditional biomarkers are not suitable for improving the accuracy of diagnosis and prognosis.
miRNAs contribute to tumorigenesis by affecting cellular processes, including the cell cycle, angiogenesis, invasion, and metastasis.Citation32–Citation34 Because miRNAs are protected from RNases and remain stable in plasma and serum, they may have a potential role as biomarkers for early cancer detection.Citation35 However, studies focusing on diagnostic or prognostic values of miRNAs in CCA are inconsistent. For example, Selaru et alCitation36 performed qRT-PCR on 18 primary CCAs and 12 normal liver specimens and found that miR-21 was 95% sensitive and 100% specific in distinguishing between CCA and normal tissues (AUC 0.995). Silakit’s reportCitation37 revealed that the sensitivity and specificity of urine-derived miR-21 were 63.6% and 71.4%, respectively, for differentiating CCA patients from healthy controls (AUC 0.682). Wang et alCitation38 suggested that high tissue-derived miR-21 expression was associated with a higher risk of intrahepatic cholangiocarcinoma (ICC) death compared to low tissue miR-21 expression and was an independent predictor of poor OS of ICC patients (HR = 3.519, 95% CI 1.411–5.702, p = 0.021). Huang et alCitation39 reported that CCA patients with high miR-21 expression had a mean 3-year OS of 15%, whereas patients with low miR-21 expression had a 33% 3-year OS (HR = 1.620, 95% CI 0.440–5.960, p = 0.046). Differences in specimen samples (ie, tissue or urine), different inclusion criteria, and different microarray techniques may explain discrepancies among these studies, but a lack of systematic evaluation complicates this conclusion. Knowledge on the diverse value of miRNAs is vague. Thus, we carried out this comprehensive and up-to-date research in a clinical context to precisely evaluate the diagnostic and prognostic accuracy of miRNAs.
In this systematic review, 12 diagnostic and 22 prognostic studies were included to study whether miRNAs are useful biomarkers for CCA. We noted that miRNAs had a relatively high diagnostic accuracy, and yielded a combined AUC of 0.88 (95% CI 0.85–0.91), with a pooled sensitivity of 0.82 and a pooled specificity of 0.83 for discriminating CCA cases. DOR, which combines the strength of sensitivity and specificity, is a useful indicator of diagnostic accuracy. In this meta-analysis, the DOR value for miRNAs was 22 (95% CI 11–43), indicating moderate diagnostic accuracy. However, the PLR and NLR were 4.8 (95% CI 3.1–7.3) and 0.22 (95% CI 0.16–0.29), respectively, suggesting that miRNAs may be insufficient in distinguishing patients with CCA because PLR > 10 and NLR < 0.1 are thresholds representing high accuracy. Subgroup and meta-regression analyses were applied if heterogeneity occurred in the diagnostic meta-analysis. We compared factors that may influence the heterogeneity and suggest that plasma specimens may be the chief sources of heterogeneity (p = 0.05). Tumor-derived circulating miRNA may have utility as a noninvasive blood biomarker, and both the sensitivity and specificity of blood-based assays (serum and plasma) exceeded 0.8, so they may have a high diagnostic value.
Elevated expression of miRNAs (miR-21, miR-26a, miR-29a, miR-181c, miR-191, miR-192, miR-200c, and miR-221) was associated with poor survival of CCA patients, and decreased expression of miRNAs (miR-34a, miR-106a, miR-203, and miR-373) was associated with a worse prognosis. Pooled HR values of OS correlated with miRNA expression for CCA patients, and this suggested that specific miRNAs are independent risk factors for prognosis and may have use for clinical decision-making. The forest plot revealed heterogeneity in this meta-analysis (I2 = 88.1%; p < 0.001), so we performed meta-regression analysis to explore the source. Results showed that the up–downregulation of miRNAs was significantly related to heterogeneity (p = 0.001) and may partly explain the heterogeneity of the OS analysis. We also evaluated the relationship between miRNA expression and RFS, DFS, and PFS. The pooled HR of PFS from the included studies was 2.31 (95% CI 1.59–3.36), indicating high expression of miRNAs might predict poor PFS for cancer patients. The included studies had low heterogeneity (I2 = 0.4%; p = 0.366), and the pooled HRs of DFS and RFS were 0.67 (0.16–2.81) and 2.68 (0.88–8.15), respectively, which showed that miRNAs can be used to monitor the therapeutic effects of radical resection or chemotherapy. However, due to limited studies, meta-regression and subgroup analyses for DFS and RFS subgroups were not performed, although there was significant heterogeneity.
Several miRNAs were confirmed to be associated with CCA patient variables, but most were assessed in one study, and only miR-21 was reported in at least three studies, suggesting it requires more investigation. For the diagnostic meta-analysis, four studies involving 210 patients and 159 healthy controls were included, and the pooled sensitivity and specificity of miR-21 were 0.85 (95% CI 0.76–0.91) and 0.92 (95% CI 0.81–0.97), respectively, with an ROC of 0.93 (95% CI 0.91–0.95). Compared with the pooled parameters of miRNAs, miR-21 had a higher diagnostic accuracy than other miRNAs and a better DOR (53, 95% CI 12–239), NLR (0.18, 95% CI 0.10–0.32), and PLR (9.4, 95% CI 3.5–25.4), indicating miR-21 was adequate to distinguish CCA patients from normal subjects. Data showed that patients with increased miR-21 had a higher risk of poor survival compared to those with low miR-21 (HR 1.88, 95% CI 1.41–2.51). There was no statistical heterogeneity among the included studies, so miR-21 may be an ideal prognostic marker for clinical decision-making. In conclusion, these findings proved that miR-21 could be a more suitable clinical biomarker with a higher capacity for discriminating CCA patients and healthy people. Exploring the sources of heterogeneity is necessary in a meta-analysis. In the present study, different measures such as sample size, ethnicity, RNA extraction, and measurement methods were used to extract miRNAs in different studies. However, there is no evidence that these variables may influence the heterogeneity. In addition, high expression of miR-21 has been found to relate with hepatocellular carcinoma (HCC).Citation40 Our study further concluded that miR-21 correlate with diagnosis and prognosis of CCA with numerous miRNAs. It may not be possible for miR-21 to distinguish HCC and CCA. Thus, clinical features and techniques such as physical examination and CT/MRI scanning may need to be combined to differentiate CCA from HCC.Citation41
miRNAs have been implicated in the pathogenesis of CCACitation42–Citation45 because upregulated miRNAs suggest an oncogenic role and downregulation of miRNAs may be suppressive, so dysregulated miRNAs are correlated with the adverse clinical features, diminished survival, and poor prognosis of CCA patients.Citation46 Selaru et alCitation36 proved that miR-21 was oncogenic by inhibiting PDCD4 and TIMP3 suppressor genes in the biliary tree. Wang et alCitation38 confirmed the functional and mechanistic links between miR-21 and tumor suppressor genes (such as PTPN14 and PTEN) in the pathogenesis of ICC, which influenced the proliferation and tumor progression in ICC. Li et alCitation47 reported that miR-221 promoted extrahepatic CCA (EHCC) invasion and metastasis by targeting PTEN and formed a positive feedback loop with the β-catenin/c-Jun signaling pathways. Moreover, results from Qiao’s studyCitation48 suggested that miR-34a inhibited invasion and migration by targeting Smad4 to suppress the epithelial–mesenchymal transition via the TGF-β/Smad signaling pathway in human EHCC. In conclusion, altered miRNA expression is a key to tumorigenesis and can be used to identify clinicopathologic features of the disease.
Our study was limited by finding miR-21 in four studies, but up to 17 miRNAs were identified to be valuable for diagnosis or prognosis, and study heterogeneity occurred. Subgroup analysis was performed to find the source of heterogeneity, but we could not fully explain it. Likely differences in patients’ baseline characteristics (ethnicity, gender, age, and tumor stage and grade), different thresholds for miRNA expression, and the way samples were prepared and preserved (ie, paraffin-fixed, formalin-fixed, freshly frozen tumors, or blood) made a difference. In addition, some HRs were calculated based on data extracted from survival curves, which might be less powerful than data obtained from articles directly.
Despite these limitations, our review had several important strengths. First, a relatively thorough systematic search was performed, and diagnostic and prognostic values of miRNAs in CCA were independently evaluated and verified. Our methods were rigorous and followed guidelines for conducting and reporting systematic reviews. Besides, further analysis and research was carried out to attest the diagnostic and prognostic roles of miR-21 expression in CCA patients.
Conclusion
miRNAs, especially miR-21, were identified to be highly correlated with CCA and could be potential and promising biomarkers in distinguishing CCA patients from healthy people; they could also contribute to predicting the progression of CCA. Furthermore, to better understand and use miRNAs as biomarkers in clinical detection, more large-scale and high-quality investigations are needed to confirm our results.
Acknowledgments
We thank the authors of all the included studies. This work was supported by a grant from the Science and Technology Project of Suzhou (SYS201764).
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRMaJZouZJemalACancer statistics, 2014CA Cancer J Clin201464192924399786
- ZhangGWLinJHQianJPZhouJIdentification of risk and prognostic factors for patients with clonorchiasis-associated intrahepatic cholangiocarcinomaAnn Surg Oncol201421113628363724781504
- SriampornSPisaniPPipitgoolVSuwanrungruangKKamsa-ardSParkinDMPrevalence of Opisthorchis viverrini infection and incidence of cholangiocarcinoma in Khon Kaen, Northeast ThailandTrop Med Int Health20049558859415117303
- ChamadolNPairojkulCKhuntikeoNHistological confirmation of periductal fibrosis from ultrasound diagnosis in cholangiocarcinoma patientsJ Hepatobiliary Pancreat Sci201421531632224420706
- MaithelSKGamblinTCKamelICorona-VillalobosCPThomasMPawlikTMMultidisciplinary approaches to intrahepatic cholangiocarcinomaCancer2013119223929394223963845
- AndersenJBThorgeirssonSSGenetic profiling of intrahepatic cholangiocarcinomaCurr Opin Gastroenterol201228326627222395571
- BlechaczBGoresGJCholangiocarcinoma: advances in pathogenesis, diagnosis, and treatmentHepatology200848130832118536057
- BrandiGVenturiMPantaleoMAErcolani G;GICOCholangiocarcinoma: current opinion on clinical practice diagnostic and therapeutic algorithms: a review of the literature and a long-standing experience of a referral centerDig Liver Dis201648323124126769568
- LeeYSDuttaAMicroRNAs in cancerAnnu Rev Pathol2009419922718817506
- CroceCMCalinGAmiRNAs, cancer, and stem cell divisionCell200512216716009126
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- MitchellPSParkinRKKrohEMCirculating microRNAs as stable blood-based markers for cancer detectionProc Natl Acad Sci U S A200810530105131051818663219
- WangLGGuJSerum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasisCancer Epidemiol2012361e61e6722018950
- KishimotoTEguchiHNaganoHPlasma miR-21 is a novel diagnostic biomarker for biliary tract cancerCancer Sci2013104121626163124118467
- SilakitRLoilomeWYongvanitPCirculating miR-192 in liver fluke-associated cholangiocarcinoma patients: a prospective prognostic indicatorJ Hepatobiliary Pancreat Sci2014211286487225131257
- LevyCLympJAnguloPGoresGJLarussoNLindorKDThe value of serum CA 19-9 in predicting cholangiocarcinomas in patients with primary sclerosing cholangitisDig Dis Sci20055091734174016133981
- ChengQFengFZhuLCirculating miR-106a is a novel prognostic and lymph node metastasis indicator for cholangiocarcinomaSci Rep201551610326534789
- WhitingPRutjesAWReitsmaJBBossuytPMKleijnenJThe development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviewsBMC Med Res Methodol200332514606960
- StroupDFBerlinJAMortonSCMeta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) groupJAMA2000283152008201210789670
- ParmarMKTorriVStewartLExtracting summary statistics to perform meta-analyses of the published literature for survival endpointsStat Med19981724281528349921604
- ReitsmaJBGlasASRutjesAWScholtenRJBossuytPMZwindermanAHBivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviewsJ Clin Epidemiol2005581098299016168343
- HigginsJPThompsonSGDeeksJJAltmanDGMeasuring inconsistency in meta-analysesBMJ2003327741455756012958120
- WuYFuXZhuXPrognostic role of systemic inflammatory response in renal cell carcinoma: a systematic review and meta-analysisJ Cancer Res Clin Oncol2011137588789620878529
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- SiricaAECholangiocarcinoma: molecular targeting strategies for chemoprevention and therapyHepatology200541151515690474
- ChuXZhaoPLvYLiuLDecreased expression of TFPI-2 correlated with increased expression of CD133 in cholangiocarcinomaInt J Clin Exp Pathol20158132833625755719
- WuTCyclooxygenase-2 and prostaglandin signaling in cholangiocarcinomaBiochim Biophys Acta20051755213515015921858
- NittaTSatoYRenXSAutophagy may promote carcinoma cell invasion and correlate with poor prognosis in cholangiocarcinomaInt J Clin Exp Pathol2014784913492125197362
- PatelTCholangiocarcinoma--controversies and challengesNat Rev Gastroenterol Hepatol20118418920021460876
- InceATYıldızKBaysalBRoles of serum and biliary CEA, CA19-9, VEGFR3, and TAC in differentiating between malignant and benign biliary obstructionsTurk J Gastroenterol2014252162169
- HuangLChenWLiangPSerum CYFRA 21-1 in biliary tract cancers: a reliable biomarker for gallbladder carcinoma and intrahepatic cholangiocarcinomaDig Dis Sci20156051273128325487191
- IorioMVFerracinMLiuCGMicroRNA gene expression deregulation in human breast cancerCancer Res200565167065707016103053
- GramantieriLFerracinMFornariFCyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinomaCancer Res200767136092609917616664
- YanaiharaNCaplenNBowmanEUnique microRNA molecular profiles in lung cancer diagnosis and prognosisCancer Cell20069318919816530703
- BernuzziFMarabitaFLleoASerum microRNAs as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinomaClin Exp Immunol20161851617126864161
- SelaruFMOlaruAVKanTMicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3Hepatology20094951595160119296468
- SilakitRLoilomeWYongvanitPUrinary microRNA-192 and microRNA-21 as potential indicators for liver fluke-associated cholangiocarcinoma risk groupParasitol Int201766447948526456596
- WangLJHeCCSuiXMiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTENOncotarget2015685932594625803229
- HuangQLiuLLiuCHMicroRNA-21 regulates the invasion and metastasis in cholangiocarcinoma and may be a potential biomarker for cancer prognosisAsian Pac J Cancer Prev201314282983423621247
- AliHEAAbdel HameedREffatHCirculating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinomaClin Res Hepatol Gastroenterol2017414e51e6228750770
- AlexanderLFHarriPLittleBMorenoCCMittalPKMagnetic resonance imaging of primary hepatic malignancies in patients with and without chronic liver disease: a pictorial reviewCureus201798e153928989828
- Peraldo NeiaCCavalloniGChiorinoGOstanoPAgliettaMLeoneFGene and microRNA modulation upon trabectedin treatment in a human intrahepatic cholangiocarcinoma paired patient derived xenograft and cell lineOncotarget2016752867668678027902465
- LoosenSHSchuellerFTrautweinCRoySRoderburgCRole of circulating microRNAs in liver diseasesWorld J Hepatol201791258659428515844
- OlaizolaPLee-LawPYArbelaizAMicroRNAs and extracellular vesicles in cholangiopathiesBiochim Biophys Acta201818644 Pt B1293130728711597
- LiZShenJChanMTVWuWKKThe role of microRNAs in intrahepatic cholangiocarcinomaJ Cell Mol Med201721117718427619971
- ChenYLiuDLiuPChenYYuHZhangQIdentification of biomarkers of intrahepatic cholangiocarcinoma via integrated analysis of mRNA and miRNA microarray dataMol Med Rep20171531051105628098904
- LiJYaoLLiGmiR-221 promotes epithelial-mesenchymal transition through targeting PTEN and forms a positive feedback loop with β-catenin/c-Jun signaling pathway in extra-hepatic cholangiocarcinomaPLoS One20151010e014116826501139
- QiaoPLiGBiWYangLYaoLWuDmicroRNA-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting Smad4 through transforming growth factor-beta/Smad pathwayBMC Cancer20151546926077733
- WangLJZhangKLZhangNSerum miR-26a as a diagnostic and prognostic biomarker in cholangiocarcinomaOncotarget2015621186311864026087181
- WangSYinJLiTUpregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinomaOncol Rep201533281982525482320
- Correa-GallegoCMaddaloDDoussotACirculating plasma levels of microRNA-21 and microRNA-221 are potential diagnostic markers for primary intrahepatic cholangiocarcinomaPLoS One2016119e016369927685844
- ChuCHChouWWangFYehCNChenTCYehTSExpression profile of microRNA-200 family in cholangiocarcinoma arising from choledochal cystJ Gastroenterol Hepatol20163151052105926479153
- ChenYLuoJTianRSunHZouSmiR-373 negatively regulates methyl-CpG-binding domain protein 2 (MBD2) in hilar cholangiocarcinomaDig Dis Sci20115661693170121086164
- LiJGaoBHuangZPrognostic significance of microRNA-203 in cholangiocarcinomaInt J Clin Exp Pathol2015889512951626464713
- ChusornPNamwatNLoilomeWOverexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasisTumour Biol20133431579158823417858
- DengYChenYIncreased expression of miR-29a and its prognostic significance in patients with cholangiocarcinomaOncol Res Treat201740312813228118648
- LiHZhouZQYangZRMicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinomaHepatology201766113615128194813
- WangJXieCPanSN-myc downstream-regulated gene 2 inhibits human cholangiocarcinoma progression and is regulated by leukemia inhibitory factor/MicroRNA-181c negative feedback pathwayHepatology20166451606162227533020
- GarajovaIBrandiGBiascoGMiR-21 expression correlates with prognosis in extrahepatic radically resected cholangiocarcinomas treated with gemcitabine-based adjuvant chemotherapyEur J Cancer201551S33S33