Abstract
Background
The independent growth factor 1 (Gfi-1) is a transcription factor essential for several diverse hematopoietic functions and developments. However, the role and molecular mechanism of Gfi-1 in the development and progression of cervical cancer remains unclear.
Purpose
The present study investigates the relation of expression of Gfi-1 with prognoses in patients with cervical cancer.
Methods
We used Western blot and reverse transcription polymerase chain reaction (RT-PCR) and the inhibition of proliferation and metastasis of cervical cancer cells in vitro.
Results
This study confirms that the expression of Gfi-1 in cervical cancer tissues was higher than that in adjacent normal tissues. The level of Gfi-1 mRNA in human cervical cancer tissues was significantly higher than that in normal tissues adjacent to cancer. Furthermore, overexpression of Gfi-1 promoted cell proliferation, colony formation, and migration of cervical cancer cells. The increased expression of Gfi-1 promotes the proliferation of cervical cancer cells targeting the tumor suppressor F-box and WD repeat domain containing 7 (FBW7). Clinically, our data suggest that overexpression of Gfi-1 is associated with poor prognosis in patients with cervical cancer. In a tumor xenograft model, knockdown of Gfi-1 inhibited the tumor growth of Hela cells in vivo.
Conclusion
Our results reveal that Gfi-1 plays an important role in cervical cancer and Gfi-1/FBW7 axis serves as a potential therapeutic target for cervical cancer.
Keywords:
Introduction
Cervical cancer is one of the leading causes of cancer deaths in women worldwide. Approximately 500,000 new cases of cervical cancer have been estimated and 280,000 people die each year.Citation1 More than 80% of cervical cancer patients and 150,000 new cases of cervical cancer in developing countriesCitation2 account for ~150,000 of China’s annual cases and ~30% of the world’s new cases. Over the past few decades, although overall incident or mortality rates have declined steadily among cervical cancer patients globally, these rates have risen significantly for patients in developing countries, including China.Citation3–Citation5 Moreover, the estimated 5-year survival rates decreased dramatically if women were diagnosed with advanced-stage cervical cancer.Citation5 The determination of tumor markers plays an important role in the early diagnosis and treatment of cervical precancerous lesions and cervical cancer, which can effectively reduce the occurrence of cervical cancer and prolong the survival time of patients with cervical cancer.Citation6
Gfi-1 is a transcription inhibitor of DNA binding. The N-terminal Snail/Gfi-1 domain is necessary for nuclear localization. In contrast, the C-terminal zinc finger domain is required for binding DNA elements to a sequence of 5′-TAAATCAC(A/T)GCA-3′.Citation7 Gfi-1 can be detected in hepatic stellate cells (HSCs), common lymphatic progenitor cells, and granule/monocyte progenitor cells, but it does not exist in common bone marrow progenitor cells and megakaryocyte/erythrocyte progenitor cells.Citation8 Studies have shown that Gfi-1 knockout mice bone marrow B- and T-cell lineages and HSCs defects indicate that Gfi-1 plays a key role in different cell lineage.Citation9 Gfi-1, a cooperating oncogene in lymphoid cells,Citation10,Citation11 unexpectedly restricts the proliferation of HSCs. Meanwhile, Gfi-1 was rapidly downregulated upon the induction of eosinophilic differentiation in late neutrophilic differentiation, but the negative effect was eosinophilic development.Citation12 Gfi-1 is an essential regulator of the ectonucleotidase expression of Th17 cells during differentiation. Th17 cells express nucleotide enzymes that inhibit the function of T cells, thereby inhibiting antitumor immunity.Citation13 Inhibiting Gfi-1 can not only cure acute lymphoblastic leukemia (ALL) in mice but also hinder the expansion of human primary cells.Citation14 Thus, Gfi-1 as a so-called “oncorequisite” factor does not play a role in the process of malignant transformation but is critically required by tumor cells for survival and progress.Citation14 Gfi-1 suppressed T-cell death by inhibiting the activation and rewriting of proapoptotic factors in G1 cell cycle checkpoints. Gfi-1 regulates the proliferation of IL-4/STAT6-dependent Th2 cells and IL-6/STAT3-mediated proliferation of antigenic stimulation.Citation15 Several targets of Gfi-1, including STAT 5B, Mcl-1, Runx2, and SOCS1, have been found in many types of cells.Citation16 These results suggest that more targets of Gfi-1 remain, encouraging further exploration to obtain deeper insights into the function of Gfi-1. However, in the occurrence and development of tumors, the definition of the mechanism of Gfi-1 expression abnormality and its regulation is still unclear; in addition, the link between Gfi-1 and cervical cancer has not been reported.
F boxes and WD repeat domains containing 7(FBW7) is apart of the substrate recognition of Skp1 Cul1 F box(SCF) that degradation of ubiquitin ligase complexes, as well as a variety of tumor suppressor proteins, including cyclin E, Notch, c-myc and c-jun, is involved in inhibiting the development of cancer.Citation17–Citation19 A recent study showed that FBXW7 is downregulated in cervical cancer and associated with risk factors of cervical cancer, such as poor grade, lymphovascular space invasion, and lymph node metastasis.Citation20 However, studies on the regulation of FBW7 upstream signaling pathways are very limited. In this study, we found that the expression of Gfi-1 was increased in cervical cancer tissues, which contributed to the adverse prognosis of the clinical patients. In addition, these findings suggest that the Gfi-1/FBW7 axis is associated with a malignant phenotype and is a potential therapeutic target for cervical cancer.
Materials and methods
Cell culture
Human embryonic kidney 293T cells and human cervical carcinoma cell lines HeLa and SiHa were purchased from the American Type Culture Collection (ATCC). All cell lines were cultured in Dulbecco’s modified eaglemedium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum (Hyclone, Logan, USA), penicillin–streptomycin (100 U/mL; Hyclone), and glutamine (2 mM; Hyclone). Cell cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C. The cells were dissociated with 0.25% trypsin and 0.02% EDTA solution and subcultured once in every 2–3 days.
Specimens
During 2013–2016, the cervical cancer tissue of 200 patients was collected in inpatient and outpatient operations at Zhongnan Hospital of Wuhan University. According to the International Federation of Gynecology and Obstetrics (FIGO) staging standards (2009) and histopathological classification, cervical cancer clinical staging, all patients provided complete clinical data and had not received chemotherapy, radiotherapy, biological treatment, or surgical treatment. After reviewing, the Department of Pathology of Wuhan University made a pathological diagnosis of each sample. The age difference of the three groups was not statistically significant.
Cell viability assay
In a 96-well plate, 2×103 cells were cultured and 20 mL of MTT solution was added at the specified time point and incubated for 4 h. The culture medium (200 mL of DMEM containing 10% fetal bovine serum) was replaced by 150 mL of DMSO, and the plate was vibrated for 10 min, and absorbance measured at 490 nm to determine the number of living cells in each well. All experiments were performed three times.
Colony formation assay
Cells were seeded in six-well plates at a density of 1×10 Citation3 cells/well and allowed to attach to the plate overnight prior to treatment. Cells were incubated for 10 days. Then, they were fixed with 4% formaldehyde and stained with crystal violet. The number of colonies with at least 50 cells was counted under a microscope at 4× magnification.
Lentivirus production and transduction of target cells
We constructed the lentiviruses vectors of overexpression Gfi-1. Gfi-1 shRNA lentivirus (TRCN0000020468; Open Bio-systems, Huntsville, AL, USA) was used to infect cells in the presence of Polybrene. Forty-eight hours later, Hela and SiHa cells were cultured in medium containing puromycin for the selection of stable clones. The FBW7 expression lentivirus was purchased from Shanghai GeneChem Co (Shanghai, China).
Coimmunoprecipitation and Western blotting
The HEK-293T cells cultured in 10 cm Petri dishes were transfected with calcium phosphate precipitation method. After 36h, the transfected cells were lysed with radio-immunoprecipitation assay (RIPA) buffer containing the complete protease inhibitor (Roche diagnosis method). The protein with the marked antibody and protein A/G beads was incubated, in a mild rotation for 4 h. Beads were washed five times with RIPA. The binding protein was eluted in buffer at 100°C for 5 min. Immunoprecipitation and protein lysis were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad Laboratories Inc., Hercules, CA, USA), and blocked with 0.5% BSA (Sigma-Aldrich Co., St Louis, MO, USA). Blots were probed with antibodies to Gfi-1 (N20; Santa Cruz Biotechnology Inc., Dallas, TX, USA), Flag (M2; Sigma-Aldrich Co.), actin (AC40; Sigma-Aldrich Co.), and hemagglutinin (SC-57592; Santa Cruz Biotechnology Inc.)
Transwell assays
The transwell assay was performed in 24-well plates. For the invasion assay, the top side of a polycarbonate filter was coated with Matrigel and placed in the upper chamber of a BioCoat Invasion Chamber (BD, Franklin Lakes, NJ, USA). Cells were resuspended in serum-free medium (2×104/200 mL), added to the top well, and incubated at 37°C for 24 h. Cells that crossed to the underside of the transwell membrane were fixed in 4% formaldehyde, then stained with 0.1% crystal violet, and counted under a microscope.
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted using the TRIzol Reagent (Sigma-Aldrich Co.) according to the manufacturer’s protocol. RNA was then treated with DNaseI (Hoffman-La Roche Ltd., Basel, Switzerland), purified by an Rneasy column (Qiagen NV, Venlo, the Netherlands), and electrophoresed in the 5′- rapid amplification of cDNA ends experiment to determine the integrity of the RNA. The complementary DNA (cDNA) was synthesized from 1 mg of total RNA using the random hexapod (progo) and superscript III reverse transcriptase (Thermo Fisher Scientific). RT-PCR was carried out on a panel of cell lines and tumor samples. The qPCR primers are as follows: 5′-GCACCGTCAAGGCT-GAGAAC-3′ and 5′-TGGTGAAGACGCCAGTGGA-3′ for GAPDH; 5′-AAAGAGTTGTTAGCGGTTCTCG-3′ and 5′-CCACATGGATACCATCAAACTG-3′ for FBW7.
Cell cycle analysis
In the cell cycle analysis, the 4×105 cells were synchronized by serum starvation for 24 h, and then re-stimulated by the addition of 10% of fetal bovine serum to re-enter the cell cycle for 9h. The cells were harvested and fixed in 75% of ethanol and kept overnight at 4°C. Cells were incubated with RNase A at 37°C for 30 min and then stained with propidium iodide (PI) at 37°C for 30 min. Cell cycle was measured using flow cytometry.
Tumor xenograft
Nude mice (BALB/c; specific pathogen-free grade; 6 weeks old, subcutaneous injection of 200 mL [5×106] cells) were used in this study. The tumor size was measured with a vernier caliper every 3 days. Tumor volumes were determined according to the following formula: A×B2/2, where A is the largest diameter and B is the perpendicular diameter. On the 30th day after the injection, the mice were sacrificed, and the tumor was withdrawn. All operations involving live mice were approved by the Animal Care and Use Committee of Wuhan University.
Statistical analyses
Means were compared using the unpaired two-tailed Student’s t-test. A P-value of <0.05 was considered statistically significant in all calculations.
Ethics approval and consent to participate
All animals were fed and used in accordance with the guidelines of the Institutional Animal Care and Use Committee at Zhongnan Hospital of Wuhan University. Ethical approval for the human tissue study was obtained from Wuhan university. Written informed consent was obtained from all participants before beginning the study.
Results
Association between Gfi-1 expression and clinicopathological characteristics
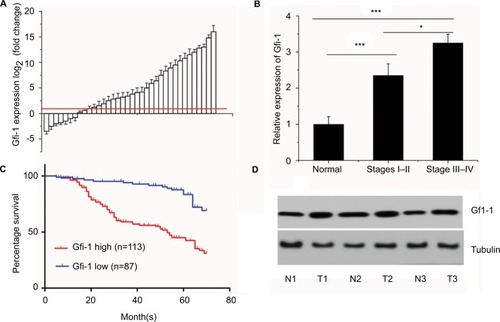
To further explore the role of Gfi-1 in cervical cancer, we conducted real-time PCR for 200 cervical cancer tissues and adjacent normal tissues in the training cohort. The Gfi-1 expression was significantly increased compared with adjacent normal tissues (). These data showed that increased expression of Gfi-1 may be a common event in cervical cancer. For clinical stage, the expression of Gfi-1 in III and IV cancer tissues was significantly higher than that in I and II cancer tissues () (P<0.01). To study the relationship between Gfi-1 expression and clinicopathological features, we analyzed its correlation in the training cohort. As shown in , the expression of Gf-1 was closely related to the degree of differentiation in cervical cancer (P=0.052), clinical stage (P<0.001), diameter of tumor (P<0.001), metastasis of lymph nodes (P<0.05) and vascular invasion (P<0.05); however, there were no significant differences between Gfi-1 expression and age, human papillomavirus infection in patients with cervical carcinoma. Furthermore, patients were divided into the following two groups: high Gfi-1 expression (n=113) and low Gfi-1 expression (n=87) groups. There was a striking negative correlation between Gfi-1 expression intensity and overall survival (OS; P<0.01) (). Western blot was conducted to investigate Gfi-1 at a protein level (). The results revealed low Gfi-1 expression in normal cervical tissues but high Gfi-1 expression in cervical squamous carcinoma tissues (P<0.001).
Figure 1 Association between Gfi-1 expression and clinicopathological characteristics.
Notes: (A) Gfi-1 expression in 200 cervical cancer tissues and adjacent normal tissues. Determination of Gfi-1 expression levels of b-actin by fluorescent quantitative PCR and b-actin standard. The red line indicates fold change of Gfi-1 equal to 2. (B) A total of 200 patients were divided into stages I, II, and III and IV. The graph shows the expression of Gfi-1 in each group. *P<0.05, ***P<0.001. (C) The OS rates of the 200 cervical cancer patients were compared with the Gfi-1 low and Gfi-1 high groups. (D) Gfi-1 protein levels were detected in tissues of normal or cervical cancer patients by Western blotting. N, normal cervical tissue. Numbers 1–6 indicate patient number. Statistical significance was determined using the log-rank test.
Abbreviations: OS, overall survival; PCR, polymerase chain reaction; T, tumor cervical tissue.
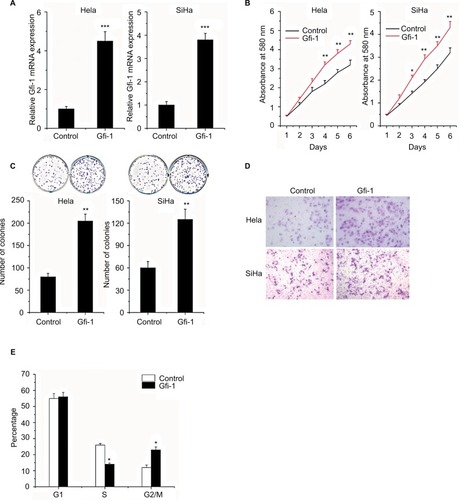
Gfi-1 promotes proliferation of human cervical cancer cells
To investigate the roles of Gfi-1 in cervical cancer cells, Gfi-1 overexpression Hela and SiHa cells were generated and the efficiency of Gfi-1 overexpression was determined by qRT-PCR assay (). Then, we performed the MTT assay to evaluate cell proliferation. Overexpression of Gfi-1 significantly increased cell proliferation in Hela and SiHa cells (). Moreover, the effect of Gfi-1 on the proliferation of cervical carcinoma cells was examined in a colony formation assay. We found that after 10 days of incubation with overexpression of Gfi-1, colony formation was significantly increased in HeLa and SiHa cells compared to the control group (). To determine Gfi-1 effects on cervical carcinoma cell migration and invasion ability, we performed wound healing and cell invasion assays. As expected, overexpression of Gfi-1 significantly promoted the migration of HeLa and SiHa cells in vitro (P<0.05) (). Since the increase in Gfi-1 results in an increased proliferation of cervical cells, cell cycle analysis is used to detect whether overexpression of Gfi-1 promotes cell cycle transitions at specific stages of the cell cycle. The results showed no difference in the percentage of overexpression of Gfi-1 cells in G1 phase, a significant decrease in S phase cells, and a significant increase in G2/M cells (). These results indicated that Gfi-1 effectively enhanced the viability of cervical carcinoma cells by promoting G2/M cell cycle transition.
Figure 2 Gfi-1 promotes proliferation and metastasis of human cervical cancer cells.
Notes: (A) The cells were transfected with Gfi-1 and then used for real-time analysis. (B) MTT assay was used to determine the cell proliferation rate of the cells with or without overexpression of Gfi-1 at the time points indicated. *P<0.05 and **P<0.01. (C) Effect of Gfi-1 overexpression on colony formation was measured in Hela and SiHa cells. The cells were seeded into six-well plates and cultured for 10 days, followed by crystal violet staining. The colony count is shown below. *P<0.05, **P<0.01, and ***P<0.001. (D) Transwell assay was performed with the cells to measure the migration rate. Three independent experiments yielded similar results. (E) Cells were transfected with Gfi-1 for 48 h, and these cells were used to perform cell cycle analysis. Statistical significance was calculated using the Student’s t-tests when only two groups were compared. *P<0.05, **P<0.01, and ***P<0.001.
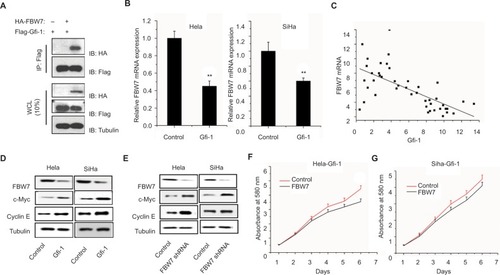
Gfi-1 target of FBW7 exerts its function by suppressing FBW7 expression
Because FBW7 functions as a ubiquitin ligase, Gfi-1 is involved in the ubiquitination and degradation of 26S proteasome in myeloid cells.Citation21 We studied whether FBW7 has a direct relationship with Gfi-1.
To this end, we transfected the HEK-293T cells into Flag–Gfi-1, with or without HA-FBW7. In the Western blot analysis, HA staining showed protein binding to FBW7 after immunoprecipitation (). This result indicated that FBW7 has a direct relationship with Gfi-1. Gfi-1 upregulation in cervical cancer of Hela cells and SiHa that reduce the mRNA expression of FBW7 (). In addition, the expression of Gfi-1 in human cervical cancer tissue was negatively correlated with mRNA expression level (). It has been shown that the key oncogenic FBW7 substrates include c-Myc, Mcl-1, and cyclin E. To better understand the mechanism of Gfi-1 involved in the proliferation of cervical cancer cells, the changes in Gfi-1-induced signal transduction pathway in SiHa and Hela cells were detected by Western blot. Interestingly, as well as Gfi-1, the FBW7-shRNA inhibits the expression of c-Myc and cyclin E, at protein levels, in Hela and SiHa cells (). Next, we studied the proliferation of Hela-Gfi-1 and SiHa-Gfi-1 and the expression of FBW7 gene and found that the expression of FBW7 was increased enough to cause the proliferation inhibition of cervical cancer cells ().
Figure 3 Gfi-1 target of FBW7 exerts its function by suppressing FBW7 expression.
Notes: (A) 293T cells were transfected with Gfi-1 either alone or together with FBW7. Whole-cell extracts were subjected to precipitation using HA antibody and examined for the indicated proteins by Western blot analysis. (B) Real-time quantitative PCR was used to detect the expression of FBW7 in two cervical cancer cell lines. (C) In cervical cancer, the negative correlation between Gfi-1 and FBW7 mRNA was observed. Data are represented as the mean ± SD, n=3. **P<0.01. (D and E) Expression of FBW7 gene and expression of c-Myc and cyclin E in cervical cancer cells were decreased by Gfi-1 overexpression. The reduction in FBW7 expression increased the expression of c-Myc and cyclin E in cervical cancer cells. (F and G) Growth of cervical cancer cells and detection by MTT.
Abbreviation: PCR, polymerase chain reaction.
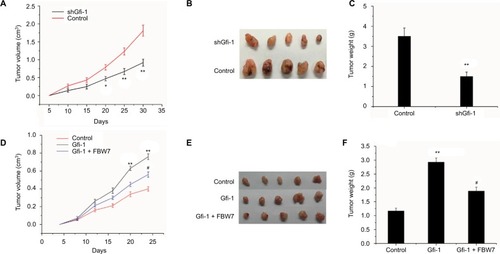
FBW7 blocks the Gfi-1-induced proliferation of Hela cells in vivo
To further explore the influence of Gfi-1 on cervical cell growth, we investigated the effects of Gfi-1 on the xenograft model. As evaluated by tumor volumes, knockdown of Gfi-1 substantially decreased Hela cells’ growth (). In addition, Hela knockdown of Gfi-1 led to a significant decrease in tumor weight compared with the control cells (). In addition, the expression of FBW7 can be increased by injecting FBW7 overexpression Hela cells and blocking the proliferation of Gfi-1 induced in xenograft models. Moreover, increased FBW7 expression was sufficient to block the Gfi-1-induced proliferation in the xenograft model by injecting FBW7 overexpression Hela cells (). These data demonstrate that Gfi-1 promotes cervical cancer tumorigenesis and FBW7 blocks the Gfi-1-induced proliferation on tumor growth in vivo.
Figure 4 FBW7 blocks the Gfi-1-induced proliferation of Hela cells in vivo.
Notes: (A and B) Tumor volume was calculated every 5 days after the injection of shGfi-1 Hela cells. Error bars indicate mean ± SD. (C) Tumor weights are represented as mean of tumor weights ± SD. *P<0.05, **P<0.01 vs control. (D and E) Tumor volume was calculated every 5 days after the injection of Gfi-1 overexpression cells and Gfi-1 and FBW7 overexpression cells. (F) Tumor weights are represented as mean of tumor weights ± SD. *P<0.05, **P<0.01 vs control group, and #P<0.05 vs Gfi-1 overexpression group.
Discussion
Cervical cancer is a malignant tumor of cervical epithelium, which is a leading gynecologic malignancy in China. The invasion and metastasis of malignant cells are mainly caused by uncontrolled proliferation of cells. Molecular biology studies show that the uncontrolled proliferation of cells is related to the abnormal expression of proto-oncogenes and tumor suppressor genes. Inactivation and mutation of tumor suppressor genes destroy the equilibrium state of cell growth. Cells lose normal control and, therefore, develop tumors or promote tumor progression.Citation22
The Gfi-1 gene encodes a 55 kDa nuclear protein that contains six C-terminal C2H2 zinc finger domains with DNA-binding properties and an important transcriptional repressor essential to the 20-amino acid barrier of the SNAG terminal domain. Gfi-1 inhibits T-cell activation by phase G1 arrest.Citation7 Gfi-1 also inhibits the regulation of apoptosis in a variety of apoptotic cells.Citation23 Meanwhile Gfi-1 counts E2F5, E2F6, Ets2, cMyc, and P21/WAF among its targets in myeloid cell lines.Citation24 Although Gfi-1 has been implicated in the pathogenesis of lymphomas, little is known about its role in cervical cancer cells. Considering the present results, we hypothesized that high Gfi-1 expression promotes the invasion and metastasis of cervical cancer. High expression of Gfi-1 is associated with risk factors for cervical cancer, such as poor grade, invasion, and metastasis.
As a tumor suppressor gene, recent studies have found that FBW7 is closely related to aberrant expression in a variety of human malignancies and is related to the prognosis of malignant tumors. FBW7 has anti-tumor effect and promotes oncogene ubiquitination and degradation, such as c-Myc, Mcl-1, cyclin E, and c-Jun, and metabolism and regulation of cell proliferation, cell differentiation, apoptosis, and other cellular processes.Citation25,Citation26 In this study, we showed that Gfi-1 directly bound the FBW7 and Gfi-1 knockdown enhanced FBW7 expression. In addition, the expression of Gfi-1 and FBW7 gene mRNA in cervical cancer tissues was negatively correlated.
Conclusion
Our results indicate that high expression of Gfi-1 is a common event in cervical cancer. Increasing Gfi-1 expression promotes cell proliferation and inhibits FBW7 ubiquitin ligase expression. This suggests that a subset of Gfi-1 was a potential therapeutic target and biomarker.
Acknowledgments
This work was supported by grants from National Natural Science Fund for Distinguished Young Scholars (no 81302274). ZL is the first author and corresponding author.
Disclosure
The authors report no conflicts of interest in this work.
References
- JemalABrayFCenterMMFerlayJWardEFormanDGlobal cancer statisticsCA Cancer J Clin2011612699021296855
- FerlayJShinHRBrayFFormanDMathersCParkinDMEstimates of worldwide burden of cancer in 2008: GLOBOCAN 2008Int J Cancer2010127122893291721351269
- Williams-BrennanLGastaldoDColeDCPaszatLSocial determinants of health associated with cervical cancer screening among women living in developing countries: a scoping reviewArch Gynecol Obstet201228661487150523011733
- BeckerSA historic and scientific review of breast cancer: the next global healthcare challengeInt J Gynaecol Obstet2015131suppl 1S36S3926433503
- Duenas-GonzalezACampbellSGlobal strategies for the treatment of early-stage and advanced cervical cancerCurr Opin Obstet Gynecol2016281111726626039
- ChoUKimHMParkHSKwonOJLeeAJeongSWNuclear expression of GS28 protein: a novel biomarker that predicts worse prognosis in cervical cancersPLoS One2016119e016262327611086
- Zweidler-MckayPAGrimesHLFlubacherMMTsichlisPNGfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressorMol Cell Biol1996168402440348754800
- ZengHYucelRKosanCKlein-HitpassLMoroyTTranscription factor Gfi-1 regulates self-renewal and engraftment of hematopoietic stem cellsEMBO J200423204116412515385956
- MoroyTKhandanpourCGrowth factor independence 1 (Gfi-1) as a regulator of lymphocyte development and activationSemin Immunol201123536837821920773
- van LohuizenMVerbeekSScheijenBWientjensEvan der GuldenHBernsAIdentification of cooperating oncogenes in E mu-myc transgenic mice by provirus taggingCell19916557377521904008
- GilksCBBearSEGrimesHLTsichlisPNProgression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger proteinMol Cell Biol1993133175917688441411
- LiuQDongFGfi-1 inhibits the expression of eosinophil major basic protein (MBP) during G-CSF-induced neutrophilic differentiationInt J Hematol201295664064722552881
- ChalminFMignotGBruchardMStat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expressionImmunity201236336237322406269
- KhandanpourCPhelanJDVassenLGrowth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemiaCancer Cell201323220021423410974
- SuzukiJMaruyamaSTamauchiHGfi-1, a transcriptional repressor, inhibits the induction of the T helper type 1 programme in activated CD4 T cellsImmunology2016147447648726749286
- LiuQBasuSQiuYTangFDongFA role of Miz-1 in Gfi-1-mediated transcriptional repression of CDKN1AOncogene201029192843285220190815
- WelckerMClurmanBEFBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiationNat Rev Cancer200882839318094723
- InuzukaHShaikSOnoyamaISCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destructionNature2011471733610410921368833
- HongSLaiminsLAThe JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiationPLoS Pathog201394e100329523593005
- XuYYuJLiuTMengFKongDLouGLoss of FBXW7 is related to the susceptibility and poor prognosis of cervical squamous carcinomaBiomarkers201621437938526954701
- ShiGMKeAWZhouJCD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinomaHepatology201052118319620578262
- McKennaNJO’MalleyBWCombinatorial control of gene expression by nuclear receptors and coregulatorsCell2002108446547411909518
- GrimesHLChanTOZweidler-McKayPATongBTsichlisPNThe Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawalMol Cell Biol19961611626362728887656
- DuanZHorwitzMTargets of the transcriptional repressor oncoprotein Gfi-1Proc Natl Acad Sci U S A2003100105932593712721361
- IbusukiMYamamotoYShinrikiSAndoYIwaseHReduced expression of ubiquitin ligase FBXW7 mRNA is associated with poor prognosis in breast cancer patientsCancer Sci2011102243944521134077
- IwatsukiMMimoriKIshiiHLoss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significanceInt J Cancer201012681828183719739118
