Abstract
Introduction
Glutathione peroxidases (GPxs) constitutes an enzyme family which has the ability to reduce free hydrogen peroxide to water and lipid hydroperoxides to their corresponding alcohols, and its main biological roles are to protect organisms from oxidative stress-induced damage. GPxs include eight members in different tissues of the body, and they play essential roles in carcinogenesis. However, the prognostic value of individual GPx in non-small cell lung cancer (NSCLC) remains elusive.
Materials and methods
In the current study, we investigated the prognostic value of GPxs in NSCLC patients through the “Kaplan–Meier plotter” database, wherein updated gene expression data and survival information from a total of 1,926 NSCLC patients are included.
Results
High expression of GPx1 mRNA was correlated with worse overall survival (OS) in adenocarcinoma patients. High expression of GPx2 mRNA was correlated with worse OS for all NSCLC patients. In contrast, high expression of GPx3 mRNA was associated with better OS for all NSCLC patients. High expression of GPx4 mRNA was significantly correlated with worsening adenocarcinoma in these patients. GPx5 mRNA high expression correlated with worsening OS for all NSCLC patients.
Discussion
The current findings of prognostic values of individual mRNA expression of GPxs in NSCLC patients indicate some GPxs may have prognostic value in NSCLC patients, and this needs further study.
Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide in both men and women.Citation1,Citation2 Non–small cell lung cancer (NSCLC) includes adenocarcinoma (ADE) and squamous cell carcinoma (SCC), and accounts for 75–80% of cases of lung cancer. A multidisciplinary approach combining the application of chemotherapy with alternative treatment modalities as well as targeted therapy have aided improvement in survival outcomes. However, success with the above treatment modalities has continually been constrained by various limitations.Citation3,Citation4 Therefore, further investigation on the mechanism of progression, as well as identification of prognostic markers, will help select patients with a higher chance of lung cancer recurrence and provide better prognosis and individualized treatment.
Glutathione peroxidases (GPxs) belong to an enzyme family which has the ability to reduce free hydrogen peroxide to water and lipid hydroperoxides to their corresponding alcohols; their main biological roles are to protect organisms from oxidative stress-induced damage.Citation5–Citation8 GPxs include eight members in different tissues of the body with different substrate specificity, and they play essential roles in the protection of the organism from oxidative damage and carcinogenesis.Citation8–Citation10 Reactive oxygen species (ROS)-induced oxidative stress from mitochondrial dysfunction or NADPH oxidase (NOX) overactivation and ectopic expression of antioxidative enzymes were involved in EGFR-mediated tumor progression and drug resistance in malignancies including NSCLC.Citation11,Citation12 However, the prognostic values of individual GPxs in NSCLC remain elusive. The “Kaplan–Meier plotter” (KM plotter) generated data from the Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/)Citation13 database. The KM plotter database was initially established using data on expression manifested by 22,277 genes in a group of 1,809 patients with breast cancer.Citation14,Citation15 Later, this database also included gene expression data and survival information from a total of 1,715 NSCLC patients.Citation16 Recently, this database has included gene expression data and survival information from patients with ovarian cancer,Citation17 gastric cancer, as well as liver cancer.Citation18 Thus, KM plotter can be utilized for the analysis of individual genes with clinical results to relapse-free survival and total survival of the patients. So far, a number of genes have been identified and/or validated by KM plotter in breast cancerCitation19–Citation27 as well as in NSCLC.Citation28 In this study, we determined the prognostic value of individual GPx in human NSCLC patients using the KM plotter database.
Materials and methods
We used an online databaseCitation14 to determine the relevance of mRNA expression of individual GPx members to relapse-free survival. The database was established using gene expression data and survival information of 1,926 NSCLC patients downloaded from Gene Expression Omnibus (GEO). Briefly, eight GPx sub-members (GPx1, GPx2, GPx3, GPx4, GPx5, GPx6, GPx7, and GPx8) were entered into the database (http://kmplot.com/analysis/index.php?p=service&cancer=lung) to obtain Kaplan–Meier survival plots in which the number-at-risk is indicated below the main plot. The certain gene mRNA expression above or below the median separates the cases into high and low expression, respectively. HR (95% CIs) and log rank p were calculated and displayed on the webpage.
Results
The GPxs family comprises a total of eight sub-members. Among all the eight GPxs isoenzymes, only GPx8 is not included in the database; GPx6 and GPx7 share the same mRNA in www.kmplot.com.
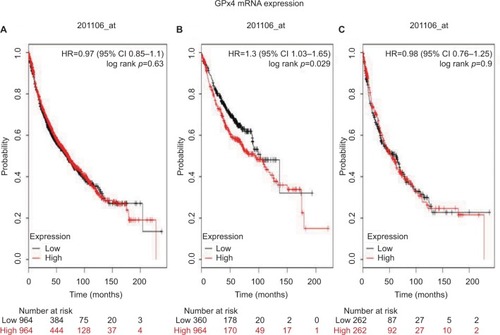
We first examined the prognostic value of GPx1 mRNA expression in www.kmplot.com. The desired Affymetrix IDs is valid: 200736_s_at (GPx1). Survival curves were plotted for all patients (n=1,926; ), for ADE (n=720; ), and for SCC (n=524; ). High expression of GPx1 mRNA was not found to be correlated with overall survival (OS) for all NSCLC patients followed for 20 years (HR 1.03 [95% CI 0.9–1.16]; p=0.7). However, high expression of GPx1 mRNA was found to be correlated to worsen OS in ADE patients (HR 1.49 [95% CI 1.18–1.89]; p=0.00083), but not in SCC patients (HR 0.81 [95% CI 0.64–1.03]; p=0.082).
Figure 1 The prognostic value of GPx1 expression according to the database of Kaplan–Meier plotter.
Notes: The desired Affymetrix ID is valid: 200736_s_at (GPx1). (A) Survival curves are plotted for all patients (n=1,926). (B) Survival curves are plotted for adenocarcinoma (n=720). (C) Survival curves are plotted for squamous cell carcinoma (n=524). Probability: overall survival.
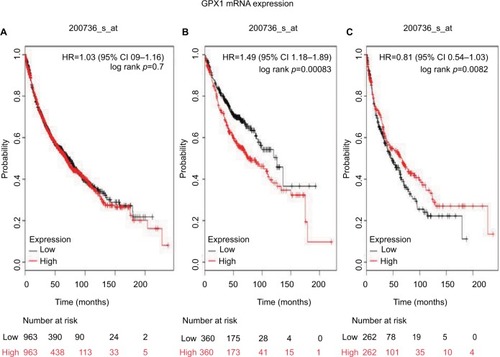
We then examined the prognostic value of GPx2 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 239595_at (GPx2). High expression of GPx2 mRNA correlated with worse OS for all NSCLC patients (HR 1.63 [95% CI 1.38–1.93]; p=5.1e-09; ). However, high expression of GPx2 mRNA was not significantly correlated with worse OS in ADE patients (HR 1.21 [95% CI 0.95–1.54]; p=0.12; ) and SCC patients (HR 1.37 [95% CI 1–1.87]; p=0.051; ).
Figure 2 The prognostic value of GPx2 expression according to the database of Kaplan–Meier plotter.
Notes: The desired Affymetrix ID is valid: 239595_at (GPx2). (A) Survival curves are plotted for all patients (n=1,145). (B) Survival curves are plotted for adenocarcinoma (n=673). (C) Survival curves are plotted for squamous cell carcinoma (n=271). Probability: overall survival.
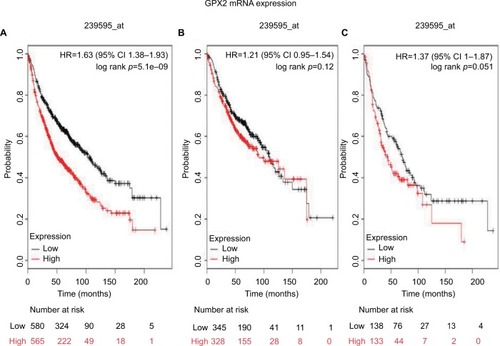
shows the prognostic value of GPx3 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 201348_at (GPx3). High expression of GPx3 mRNA correlated with better OS for all NSCLC patients (HR 0.76 [95% CI 0.67–0.86]; p=2e-05; ). However, high expression of GPx3 mRNA was not significantly correlated with better OS in ADE patients (HR 0.85 [95% CI 0.67–1.07]; p=0.16; ) and SCC patients (HR 0.87 [95% CI 0.69–1.11]; p=0.27; ).
Figure 3 The prognostic value of GPx3 expression according to the database of Kaplan–Meier plotter.
Notes: The desired Affymetrix IDs is valid: 201348_at (GPx3). (A) Survival curves are plotted for all patients (n=1,926). (B) Survival curves are plotted for adenocarcinoma (n=720). (C) Survival curves are plotted for squamous cell carcinoma (n=524). Probability: overall survival.
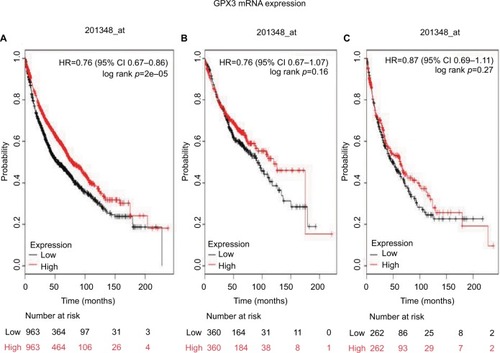
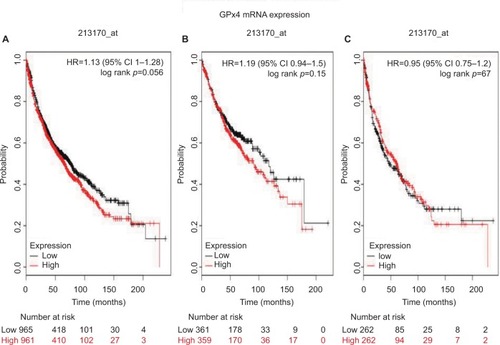
shows the prognostic value of GPx4 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 201106_at (GPx4). The curves show that high expression of GPx4 mRNA above or below the median do not separate the cases into significantly different prognostic groups among all NSCLC patients (HR 0.97 [95% CI 0.85–1.1]; p=0.63; ). However, high expression of GPx4 mRNA was significantly correlated with worse OS in ADE patients (HR 1.3 [95% CI 1.03–1.65]; p=0.029; ), but not in SCC patients (HR 0.98 [95% CI 0.78–1.25]; p=0.9; ).
Figure 4 The prognostic value of GPx4 expression according to the database from Kaplan Meier plotter.
Notes: The desired Affymetrix ID is valid: 201106_at (GPx4). (A) Survival curves are plotted for all patients (n=1,926). (B) Survival curves are plotted for adenocarcinoma (n=720). (C) Survival curves are plotted for squamous cell carcinoma (n=524). Probability: overall survival.
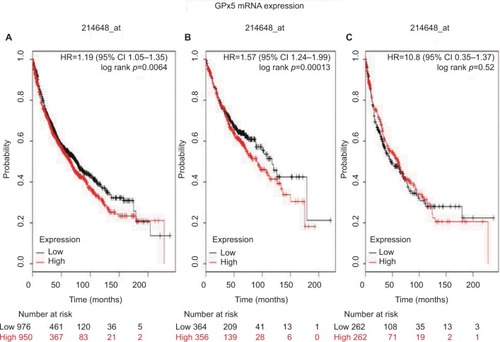
shows the prognostic value of GPx5 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 214648_at (GPx5). GPx5 mRNA high expression was found to be correlated with worse OS for all NSCLC patients (HR 1.19 [95% CI 1.05–1.35]; p=0.0064; ). In addition, high expression of GPx5 mRNA was found to be correlated with worse OS in ADE patients (HR 1.57 [95% CI 1.24–1.99]; p=0.00013; ), but not in SCC patients (HR 1.08 [95% CI 0.85–1.37]; p=0.52; ).
Figure 5 The prognostic value of GPx5 expression according to the database of Kaplan–Meier plotter.
Notes: The desired Affymetrix ID is valid: 214648_at (GPx5). (A) Survival curves are plotted for all patients (n=1,926). (B) Survival curves are plotted for adenocarcinoma (n=720). (C) Survival curves are plotted for squamous cell carcinoma (n=524). Probability: overall survival.
shows the prognostic value of GPx6 or GPx7 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 213170_at (GPx6 or7). The curves show that high expression of GPx6/7 mRNA above or below the median do not separate the cases into significantly different prognostic groups in all NSCLC patients (HR 1.13 [95% CI 1–1.28]; p=0.056; ), in ADE patients (HR 1.19 [95% CI 0.94–1.5]; p=0.15; ), as well as in SCC patients (HR 0.95 [95% CI 0.75–1.2]; p=0.67; ).
Figure 6 The prognostic value of GPx6/7 expression according to the database of Kaplan–Meier plotter.
Notes: The desired Affymetrix ID is valid: 213170_at (GPx6/7). (A) Survival curves are plotted for all patients (n=1,926). (B) Survival curves are plotted for adenocarcinoma (n=720). (C) Survival curves are plotted for squamous cell carcinoma (n=524). Probability: overall survival.
To further determine the correlation of individual GPx with other clinicopathological features, we determined their correlation with the smoking status (), clinical stages (), and chemotherapy () of NSCLC patients. As shown in , high expression of GPx5, GPx6, or GPx7 mRNA was correlated with worse OS in never-smoked NSCLC patients. From , it is apparent that GPx2, GPx3, GPx6, or GPx7 are significantly associated with clinical stages of NSCLC patients. From the data in , it is evident that none of the GPxs are significantly associated with NSCLC patients, with or without chemotherapy, probably due to the relatively limited number of patients.
Table 1 Correlation of GPx mRNA expression with smoking status of NSCLC patients
Table 2 Correlation of GPx mRNA expression with clinical stages of NSCLC patients
Table 3 Correlation of GPx mRNA expression with chemotherapy of NSCLC patients
Discussion
Changes in GPx levels in several types of tumor have been reported. GPx1 was reported to prevent oxidative DNA mutations and, thus, GPx1 may prevent tumorigenesis.Citation29 Overexpressed GPx1 reduced tumor growth, which indicates that it has a role in protecting against tumorigenesis.Citation30 The dual role of GPx2 in tumorigenesis has been reviewed recently.Citation9,Citation31 Overexpression of GPx2 was observed in several tumors,Citation32,Citation33 including lung cancer,Citation34 indicating that GPx2 may be an oncogene. GPx3 is considered to be a novel tumor suppressor, as hypermethylation of GPx3 was detected in tumor samples from patients with Barrett’s esophagusCitation35–Citation37 as well as in endometrialCitation38 and prostate cancers,Citation39 and downregulation of GPx3 was generally correlated with worse prognosis. Moreover, GPx4 is considered to be a tumor suppressor, because it is downregulated in pancreaticCitation30 and breast cancers.Citation40 In addition, GPx4 overexpression reduced fibrosarcoma cell growth.Citation41 So far, there are no reports on the role of GPx5, -6, -7, and -8 in tumorigenesis. With the exception of GPx3, all GPx members were reported to be associated with the prognosis of tumor patients. For example, increased GPx1 expression was significantly associated with poor prognosis in oral SCCCitation42 and laryngeal SCC.Citation43 In contrast, loss of GPx1 expression was associated with aggressiveness and poor survival in patients with gastric cancer.Citation43 Furthermore, GPx2 overexpression was correlated with poor prognosis in patients with hepatocellular carcinomaCitation45 and gastric cancer.Citation46 In contrast, loss of GPx2 expression was associated with patients who had esophageal SCC.Citation47 The distinct different prognostic values of GPx1 and GPx2 in different types of cancer patients have not been well explained so far.Citation48
Several reports exist about the liver changes associated with mRNA expression of GPxs in NSCLC cell lines and/or tumor tissues;Citation49–Citation51 however, the prognostic value of mRNA expression of GPxs in NSCLC patients has not been reported. KM plotter is a widely used database containing gene expression data and survival information.Citation17 Until now, a number of genes, such as ALDH1, GLI1, ITIH5, CK2, RAI14, and GREB1, have been identified and validated by KM plotter in lung,Citation16,Citation28,Citation52,Citation53 breast,Citation19–Citation28,Citation54 gastric,Citation55,Citation56 and ovarian cancers.Citation28,Citation57,Citation58 Using KM plotter, we determined the prognostic value of individual GPx mRNA in NSCLC patients. High expression of GPx1 mRNA was not correlated with OS for all NSCLC patients followed for 20 years. However, high expression of GPx1 mRNA was correlated with worse OS in ADE patients (HR 1.49 [95% CI 1.18–1.89]; p=0.00083). High expression of GPx2 mRNA correlated with worse OS for all NSCLC patients (HR 1.63 [95% CI 1.38–1.93]; p=5.1e-09). In contrast, high expression of GPx3 mRNA correlated with better OS for all NSCLC patients (HR 0.76 [95% CI 0.67–0.86]; p=2e-05). High expression of GPx4 mRNA was significantly correlated with worse OS in ADE patients (HR 1.3 [95% CI 1.03–1.65]; p=0.029). High expression of GPx5 mRNA correlated with worse OS for all NSCLC patients (HR 1.19 [95% CI 1.05–1.35]; p=0.0064). High expression of GPx6/7mRNA was not correlated with OS for all NSCLC patients. GPx8, as a novel member belonging to the GPx family, has been identified in a phylogenetic analysis in amphibia and mammalia.Citation59 GPx8 is a membrane protein, lung-abundant enzyme, and is detected in the endoplasmic reticulum.Citation60,Citation61 However, little is known about its role. In addition, individual GPx may interact among themselves and finally affect the prognosis of NSCLC patients. However, the KM plotter cannot be used to analysis the impact between the various isoforms of GPx. Moreover, the KM plotter cannot be used to analyze the correlation between isoenzyme expression.
Nicotine can regulate a number of biological functions such as cell proliferation, invasion, inflammation, apoptosis, and angiogenesis.Citation62 Through inducing the secretion of stem cell factors, nicotine is able to regulate the growth and metastasis of NSCLC.Citation63 There is no evidence about the direct correlation between nicotine and GPx expression in NSCLC. However, nicotine treatment may significantly impact the expression of GPx1 in peripheral blood lymphocytes,Citation64 rat kidneys,Citation65 as well as in a novel cell line – Danio rerio gill (DrG) – derived from the gill tissue of zebrafish.Citation66 Furthermore, nicotine treatment was reported to significantly impact the expression of GPx4, GPx6, and GPx7 in rat brain.Citation67 In this report, we observed that high expression of GPx5, GPx6, or GPx7 mRNA correlated with worse OS in never-smoked NSCLC patients.
There are a number of reports about the prognostic values of GPxs in tumor patients. For example, loss of GPx1 expression was associated with tumor aggressiveness and poor survival in patients with gastric cancer.Citation43 GPx2 overexpression indicates poor prognosis in patients with hepatocellular carcinoma,Citation45 gastric cancer,Citation46 and urothelial carcinomas of the upper urinary tract and urinary bladder.Citation68 However, patients with prostate cancer with high GPx2 expression on the biopsy specimen had significantly lower prostate-specific antigen (PSA), recurrence-free survival, and OS than those without any GPx2 expression.Citation69 Downregulation of GPx3 significantly correlated with poor prognosis in hepatocellular carcinoma,Citation70 cervical cancer,Citation71 and gallbladder cancer.Citation72 GPx3 is considered to be a novel tumor suppressor, because hypermethylation of the GPx3 promoter was detected in tumor samples from patients with Barrett’s esophagusCitation35–Citation37 as well as endometrialCitation38 and prostate cancers,Citation39 and downregulation of GPx3 was generally associated with a poor prognosis. However, so far, no prognostic significance of GPxs protein expression in NSCLC patients has been reported. The current findings of prognostic values of individual mRNA expression of GPxs in NSCLC patients indicate some members of the GPx family may also have prognostic values in NSCLC patients, and this needs further study.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (grant no. 81300064).
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRNaishadhamDJemalACancer statistics, 2012CA Cancer J Clin2012621102922237781
- GuoPHuangZLYuPLiKTrends in cancer mortality in China: an updateAnn Oncol201223102755276222492700
- LazzariCKarachaliouNBulottaACombination of immunotherapy with chemotherapy and radiotherapy in lung cancer: is this the beginning of the end for cancer?Ther Adv Med Oncol201810 1758835918762094
- JonesCMBrunelliACallisterMEFranksKNMultimodality treatment of advanced non-small cell lung cancer: where are we with the evidence?Curr Surg Rep201862529456881
- LubosELoscalzoJHandyDEGlutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunitiesAntioxid Redox Signal20111571957199721087145
- BattinEEBrumaghimJLAntioxidant activity of sulfur and selenium: a review of reactive oxygen species scavenging, glutathione peroxidase, and metal-binding antioxidant mechanismsCell Biochem Biophys200955112319548119
- ToppoSFlohéLUrsiniFVaninSMaiorinoMCatalytic mechanisms and specificities of glutathione peroxidases: variations of a basic schemeBiochim Biophys Acta20091790111486150019376195
- Brigelius-FlohéRKippAGlutathione peroxidases in different stages of carcinogenesisBiochim Biophys Acta20091790111555156819289149
- Brigelius-FlohéRKippAPPhysiological functions of GPx2 and its role in inflammation-triggered carcinogenesisAnn N Y Acad Sci20121259192522758632
- ZhuoPDiamondAMMolecular mechanisms by which selenoproteins affect cancer risk and progressionBiochim Biophys Acta20091790111546155419289153
- WengMSChangJHHungWYYangYCChienMHThe interplay of reactive oxygen species and the epidermal growth factor receptor in tumor progression and drug resistanceJ Exp Clin Cancer Res20183716129548337
- KumariSBadanaAKGMMGSMallaRReactive oxygen species: a key constituent in cancer survivalBiomark Insights201813 1177271918755391
- EdgarRDomrachevMLashAEGene Expression Omnibus: NCBI gene expression and hybridization array data repositoryNucleic Acids Res20021130120721011752295
- GyörffyBLanczkyAEklundACAn online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patientsBreast Cancer Res Treat2010123372573120020197
- GyörffyBBenkeZLánczkyARecurrenceOnline: an online analysis tool to determine breast cancer recurrence and hormone receptor status using microarray dataBreast Cancer Res Treat201213231025103421773767
- GyörffyBSurowiakPBudcziesJLánczkyAOnline survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancerPLoS One2013812e8224124367507
- GyorffyBLánczkyASzállásiZImplementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patientsEndocr Relat Cancer201219219720822277193
- SzászAMLánczkyANagyÁCross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patientsOncotarget2016731493224933327384994
- LiuMWangGGomez-FernandezCRGuoSGREB1 functions as a growth promoter and is modulated by IL6/STAT3 in breast cancerPLoS One2012710e4641023056300
- TilghmanSLTownleyIZhongQProteomic signatures of acquired letrozole resistance in breast cancer: suppressed estrogen signaling and increased cell motility and invasivenessMol Cell Proteomics20131292440245523704778
- ZhouCZhongQRhodesLVProteomic analysis of acquired tamoxifen resistance in MCF-7 cells reveals expression signatures associated with enhanced migrationBreast Cancer Res2012142R4522417809
- MaciejczykASzelachowskaJCzapigaBElevated BUBR1 expression is associated with poor survival in early breast cancer patients: 15-year follow-up analysisJ Histochem Cytochem201361533033923392733
- MaciejczykAŁackoAEkiertMElevated nuclear S100P expression is associated with poor survival in early breast cancer patientsHistol Histopathol201328451352423364898
- MaciejczykAJagodaEWysockaTABCC2 (MRP2, cMOAT) localized in the nuclear envelope of breast carcinoma cells correlates with poor clinical outcomePathol Oncol Res201218233134221986666
- AdamMANew prognostic factors in breast cancerAdv Clin Exp Med201322151523468257
- IvanovaLZandbergaESilinaKPrognostic relevance of carbonic anhydrase IX expression is distinct in various subtypes of breast cancer and its silencing suppresses self-renewal capacity of breast cancer cellsCancer Chemother Pharmacol201575223524625422154
- WuSXueWHuangXDistinct prognostic values of ALDH1 isoenzymes in breast cancerTumour Biol20151313
- OrtegaCESeidnerYDominguezIMining CK2 in cancerPLoS One2014912e11560925541719
- BaligaMSWangHZhuoPSchwartzJLDiamondAMSelenium and GPx-1 overexpression protect mammalian cells against UV-induced DNA damageBiol Trace Elem Res2007115322724217625244
- LiuJDuJZhangYSuppression of the malignant phenotype in pancreatic cancer by overexpression of phospholipid hydroperoxide glutathione peroxidaseHum Gene Ther200617110511616409129
- Brigelius-FlohéRMaiorinoMGlutathione peroxidasesBiochim Biophys Acta2013183053289330323201771
- Al-TaieOHUceylerNEubnerUExpression profiling and genetic alterations of the selenoproteins GI-GPx and SePP in colorectal carcinogenesisNutr Cancer200448161415203372
- ChiuSTHsiehFJChenSWChenCLShuHFLiHClinicopathologic correlation of up-regulated genes identified using cDNA microarray and real-time reverse transcription-PCR in human colorectal cancerCancer Epidemiol Biomarkers Prev200514243744315734970
- WoenckhausMKlein-HitpassLGrepmeierUSmoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancersJ Pathol2006210219220416915569
- LeeOJSchneider-StockRMcChesneyPAHypermethylation and loss of expression of glutathione peroxidase-3 in Barrett’s tumorigenesisNeoplasia20057985486116229808
- PengDFRazviMChenHDNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinomaGut200958151518664505
- HeYWangYLiPZhuSWangJZhangSIdentification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinomaDig Dis Sci201156368168820725785
- FalckEKarlssonSCarlssonJHeleniusGKarlssonMKlinga-LevanKLoss of glutathione peroxidase 3 expression is correlated with epigenetic mechanisms in endometrial adenocarcinomaCancer Cell Int2010104621106063
- YuYPYuGTsengGGlutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasisCancer Res200767178043805017804715
- CejasPGarcía-CabezasMACasadoEPhospholipid hydroperoxide glutathione peroxidase (PHGPx) expression is downregulated in poorly differentiated breast invasive ductal carcinomaFree Radic Res200741668168717516241
- HeirmanIGinnebergeDBrigelius-FlohéRBlocking tumor cell eicosanoid synthesis by GP x 4 impedes tumor growth and malignancyFree Radic Biol Med200640228529416413410
- LeeJRRohJLLeeSMOverexpression of glutathione peroxidase 1 predicts poor prognosis in oral squamous cell carcinomaJ Cancer Res Clin Oncol2017143112257226528653098
- ZhangQXuHYouYZhangJChenRHigh Gpx1 expression predicts poor survival in laryngeal squamous cell carcinomaAuris Nasus Larynx2018451131928641905
- MinSYKimHSJungEJJungEJJeeCDKimWHPrognostic significance of glutathione peroxidase 1 (GPX1) down-regulation and correlation with aberrant promoter methylation in human gastric cancerAnticancer Res20123283169317522843889
- LiuTKanXFMaCGPX2 overexpression indicates poor prognosis in patients with hepatocellular carcinomaTumour Biol2017396 1010428317700410
- LiuDSunLTongJChenXLiHZhangQPrognostic significance of glutathione peroxidase 2 in gastric carcinomaTumour Biol2017396 1010428317701443
- LeiZTianDZhangCZhaoSSuMClinicopathological and prognostic significance of GPX2 protein expression in esophageal squamous cell carcinomaBMC Cancer20161641027388201
- JiaoYWangYGuoSWangGGlutathione peroxidases as oncotargetsOncotarget2017845800938010229108391
- GresnerPGromadzinskaJJablonskaEKaczmarskiJWasowiczWExpression of selenoprotein-coding genes SEPP1, SEP15 and hGPX1 in non-small cell lung cancerLung Cancer2009651344019058871
- HuYRahlfsSMersch-SundermannVBeckerKResveratrol modulates mRNA transcripts of genes related to redox metabolism and cell proliferation in non-small-cell lung carcinoma cellsBiol Chem2007388220721917261084
- RomanowskaMKikawaKDFieldsJREffects of selenium supplementation on expression of glutathione peroxidase isoforms in cultured human lung adenocarcinoma cell linesLung Cancer2007551354217052796
- YouQGuoHXuDDistinct prognostic values and potential drug targets of ALDH1 isoenzymes in non-small-cell lung cancerDrug Des Devel Ther2015950875097
- DötschMMKlotenVSchlensogMLow expression of ITIH5 in adenocarcinoma of the lung is associated with unfavorable patients’ outcomeEpigenetics2015101090391226252352
- HongCQZhangFYouYJElevated C1orf63 expression is correlated with CDK10 and predicts better outcome for advanced breast cancers: a retrospective studyBMC Cancer20151554826209438
- YuTJiaWAnQCaoXXiaoGBioinformatic analysis of GLI1 and related signaling pathways in chemosensitivity of gastric cancerMed Sci Monit2018241847185529596399
- HeXYZhaoJChenZQJinRLiuCYHigh expression of retinoic acid induced 14 (RAI14) in gastric cancer and its prognostic valueMed Sci Monit2018242244225129654694
- KamieniakMMRicoDMilneRLDeletion at 6q24.2-26 predicts longer survival of high-grade serous epithelial ovarian cancer patientsMol Oncol20159242243625454820
- GayarreJKamieniakMMCazorla-JiménezAThe NER-related gene GTF2H5 predicts survival in high-grade serous ovarian cancer patientsJ Gynecol Oncol2016271e726463438
- ToppoSVaninSBoselloVTosattoSCEvolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamilyAntioxid Redox Signal20081091501151418498225
- NguyenVDSaaranenMJKaralaARTwo endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formationJ Mol Biol2011406350351521215271
- YamadaYLimmonGVZhengDMajor shifts in the spatio-temporal distribution of lung antioxidant enzymes during influenza pneumoniaPLoS One201272e3149422355371
- CardinaleANastrucciCCesarioARussoPNicotine: specific role in angiogenesis, proliferation and apoptosisCrit Rev Toxicol2012421688922050423
- PerumalDPillaiSNguyenJSchaalCCoppolaDChellappanSPNicotinic acetylcholine receptors induce c-Kit ligand/Stem Cell Factor and promote stemness in an ARRB1/β-arrestin-1 dependent manner in NSCLCOncotarget2014521104861050225401222
- SudheerARMuthukumaranSKalpanaCSrinivasanMMenonVPProtective effect of ferulic acid on nicotine-induced DNA damage and cellular changes in cultured rat peripheral blood lymphocytes: a comparison with N-acetylcysteineToxicol In Vitro200721457658517222527
- AkkoyunHTKaradenizAInvestigation of the protective effect of ellagic acid for preventing kidney injury in rats exposed to nicotine during the fetal periodBiotech Histochem201691210811526529089
- Nathiga NambiKSAbdul MajeedSTajuGSivasubbuSSarath BabuVSahul HameedASEffects of nicotine on zebrafish: a comparative response between a newly established gill cell line and whole gillsComp Biochem Physiol C Toxicol Pharmacol2017195687728257922
- SongGNesilTCaoJYangZChangSLLiMDNicotine mediates expression of genes related to antioxidant capacity and oxidative stress response in HIV-1 transgenic rat brainJ Neurovirol201622111412426306689
- ChangIWLinVCHungCHGPX2 underexpression indicates poor prognosis in patients with urothelial carcinomas of the upper urinary tract and urinary bladderWorld J Urol201533111777178925813210
- NaikiTNaiki-ItoAAsamotoMGPX2 overexpression is involved in cell proliferation and prognosis of castration-resistant prostate cancerCarcinogenesis20143591962196724562575
- QiXNgKTLianQZClinical significance and therapeutic value of glutathione peroxidase 3 (GPx3) in hepatocellular carcinomaOncotarget2014522111031112025333265
- ZhangXZhengZYingjiSDownregulation of glutathione peroxidase 3 is associated with lymph node metastasis and prognosis in cervical cancerOncol Rep20143162587259224788695
- YangZLYangLZouQPositive ALDH1A3 and negative GPX3 expressions are biomarkers for poor prognosis of gallbladder cancerDis Markers201335316317224167362
