Abstract
Background
Centromere protein F (CENPF) is a key component of the kinetochore complex and plays a crucial role in chromosome segregation and cell cycle progression. Recent work suggests that CENPF upregulation is linked to aggressive tumor features in a variety of malignancies including prostate cancer.
Materials and methods
Using a highly annotated tissue microarray, we analyzed CENPF protein expression from a cohort of 8,298 prostatectomized patients by immunohistochemistry to study its effect on prostate-specific antigen recurrence-free survival.
Results
CENPF overexpression was found in 53% of cancers, and was linked to higher Gleason grade, advanced pathological tumor stage, accelerated cell proliferation, and lymph node metastasis (p<0.0001, each). A comparison with other key molecular features accessible through the microarray revealed strong associations between CENPF overexpression and presence of erythroblast transformation-specific (ETS)-related gene (ERG) fusion as well as phosphatase and tensin homolog deletion (p<0.0001, each). CENPF overexpression was linked to early biochemical recurrence. A subset analysis revealed that this was driven by the ERG-negative subset (p<0.0001). This was independent of established preoperative and postoperative prognostic parameters in multivariate analyses.
Conclusion
The results of our study identify CENPF overexpression as an important mechanism and a potential biomarker for prostate cancer aggressiveness.
Introduction
In Western societies, the most prevalent cancer in men is prostate cancer.Citation1 The majority of prostate cancers are indolent. Only a small subset is highly aggressive and requires treatment.Citation2,Citation3 Gleason grade and tumor extent on biopsies, prostate-specific antigen (PSA) levels, and clinical stage are established preoperative prognostic parameters. These parameters are statistically powerful in retrospective analysis but insufficient for optimal individual treatment decisions. The hope is that a further understanding of disease biology will identify additional clinically applicable molecular markers for more reliable predictions of prostate cancer aggressiveness.
Centromere protein F (CENPF, also known as mitosin) is a microtubule-associated protein involved in mitosis and cell differentiation.Citation4 CENPF becomes upregulated during the G2/M phase and accumulates to the kinetochore complex, facilitating microtubule attachment and chromosome segregation.Citation5,Citation6 In addition, CENPF is essential – and probably rate limiting – for cell proliferation as it interacts with key cell cycle checkpoint proteins, such as the retinoblastoma proteinCitation4 and the Bub1 kinase,Citation7 or late telophase proteins including syntaxin 4Citation8 and SNAP25.Citation9 CENPF upregulation has been observed in breast cancer,Citation11 nasopharyngeal cancer,Citation12 hepatocellular carcinoma,Citation13 esophageal squamous cell carcinoma,Citation14 gastrointestinal stromal tumors,Citation15 and non-Hodgkin’s lymphoma,Citation16 and in some cases it is associated with aggressive tumor phenotype and poor survival.Citation11,Citation12,Citation14
Aytes et al showed that in prostate cancer cosilencing of the fork head box protein M1 (FOXM1) and CENPF abrogates tumor cell growth.Citation10 Zhuo et al described higher CENPF expression in 99 cancer samples compared with normal tissue and suggested an association with unfavorable tumor phenotype and poor prognosis.Citation17 Similar results were mentioned for the outcome of 821 patients with prostate cancers.Citation10 These promising data encouraged us to interrogate the prognostic impact of CENPF protein expression in a large set of prostate cancers. Therefore, we performed immunohistochemical analysis of CENPF protein expression on a panel of >11,000 prostate cancer specimens associated with follow-up information and an attached molecular database.
Materials and methods
Patients
The 12,427 radical prostatectomy specimens were from patients who had surgery between 1992 and 2014 at the Department of Urology and the Martini Clinic at the University Medical Center Hamburg-Eppendorf. The specimens were analyzed according to a standard procedure.Citation18 Tumor stage, Gleason grade, nodal status, and status of the resection margin were retrieved from the patients’ records. Quantitative Gleason grading was performed using the percentage of Gleason 4 patterns.Citation19 Follow-up data were available for 11,665 patients with a median follow-up of 50 months (range: 1–264 months; ). PSA values were measured following surgery, and PSA recurrence was defined as a postoperative PSA of 0.2 ng/mL and increasing in subsequent measurements. The tissue microarray manufacturing process was described in detail earlier.Citation20 In short, a single 0.6 mm core was taken from a representative tissue block. For internal control, various control tissues and normal prostate tissue were included. The molecular database attached to this tissue microarray contains results on erythroblast transformation-specific (ETS)-related gene (ERG) protein expression,Citation21 ERG rearrangement analysis by fluorescence in situ hybridization (FISH)Citation22 and deletion status of 5q21 (chromodomain-helicase-DNA-binding protein 1 [CHD1]),Citation23 6q15 (mitogen-activated protein kinase kinase kinase 7 [MAP3K7]),Citation24 10q23 (phosphatase and tensin homolog [PTEN]),Citation25 and 3p13 (fork head box protein P1 [FOXP1])Citation26 cancers. The use of leftover archived diagnostic tissues for the manufacturing of tissue microarrays and their analysis in conjunction with anonymized patient data for research purposes was approved by local laws (HmbKHG, §12,1) and by the local ethics committee (Ethics commission Hamburg, WF-049/09 and PV3652). All work was carried out in compliance with the Helsinki Declaration.
Table 1 Pathological and clinical data of the arrayed prostate cancers
Immunohistochemistry (IHC)
Freshly cut tissue microarray sections were stained the same day and in a single run. Slides were deparaffinized and exposed to heat-induced antigen retrieval for 5 minutes at 121°C in pH 9.0 antigen retrieval buffer. Primary antibody specific for CENPF (mouse monoclonal antibody ab90, dilution 1:1350; Abcam, Cambridge, UK) was applied at 37°C for 60 minutes. Bound antibody was visualized with the EnVision Kit (Dako, Glostrup, Denmark). Since CENPF typically shows nuclear and cytoplasmic staining in 100% of the tumor cells of a tissue spot, we recorded only the staining intensity as negative, weak, moderate, and strong staining.
Statistics
JMP 12.0 software (SAS Institute Inc., NC, USA) was used. Contingency tables were calculated to study the association between CENPF expression and other clinicopathological variables, and the likelihood test was used to find significant relationships. Analysis of variance and F test were applied to find associations between CENPF expression and tumor cell proliferation as measured by the Ki67-labeling index. Kaplan–Meier curves were generated using biochemical (PSA) recurrence as the clinical endpoint. The log-rank test was applied to test the significance of differences between stratified survival functions.
Results
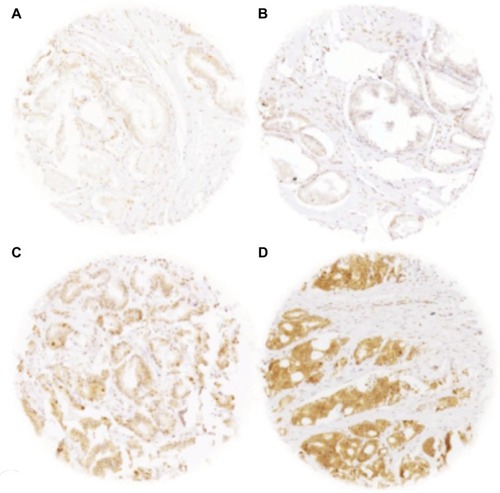
A total of 9,055 (73%) of the 12,427 arrayed tumor samples displayed interpretable CENPF staining. Noninformative cases (27%) either had no tissue or an absence of unequivocal cancer tissue in the microarray spot. Normal prostatic glands showed negative to weak cytoplasmic CENPF staining in luminal and basal cells. Comparable staining was also found in stromal cells. In cancer cells, CENPF-positive staining was seen in 8,066 of our 9,055 (89%) interpretable prostate cancers and was considered weak in 36.6%, moderate in 34.9%, and strong in 17.6% of cancers. Representative images of CENPF staining in normal and cancerous prostate samples are given in .
Figure 1 Representative pictures of CENPF staining in (A) normal prostate glands (negative/weak) and in prostate cancer with (B) negative/weak, (C) moderate, and (D) strong staining intensity. Spot size 600 µm, 100× magnification.
Abbreviation: CENPF, centromere protein F.
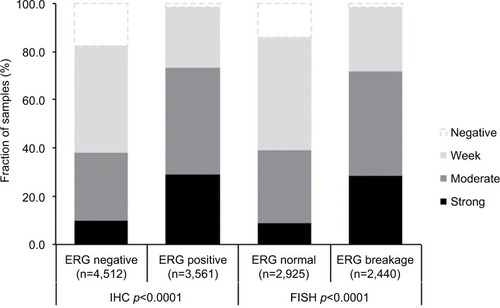
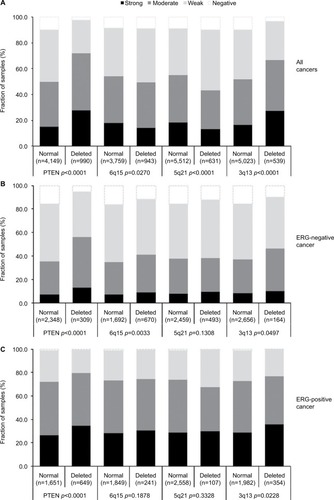
Association with TMPRSS2:ERG fusion status and ERG protein expression
To evaluate whether CENPF expression is associated with ERG status in prostate cancers, we used data from previous studies.Citation21,Citation22. Data on transmembrane protease, serine 2 (TMPRSS2):ERG fusion status obtained by FISH were available from 5,365 tumors and by IHC from 8,073 tumors with evaluable CENPF staining. Data on both ERG-FISH and IHC were available from 5,198 cancers, and an identical result (ERG-IHC positive and break by FISH or ERG-IHC negative and no break by FISH) was found in 4,975 of 5,198 (95.7%) cancers. Increased (ie, moderate to strong) CENPF staining was more frequent in TMPRSS2:ERG rearranged (72.05%) and ERG-positive prostate cancers (73.46%) than in ERG-negative tumors by FISH (39.11%) or IHC (38.3%, p<0.0001 each, ). It is of note that ERG expression was not associated with the prognosis as indicated by identical Kaplan–Meier curves for PSA recurrence-free survival in the ERG-negative and ERG-positive subset (Figure S1).
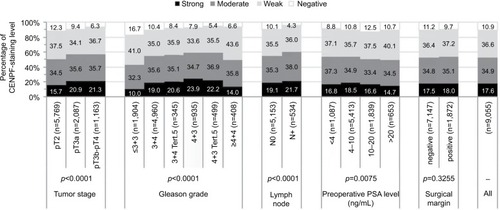
Association with tumor phenotype
Increasing CENPF staining was linked to advanced pathological tumor stage, higher Gleason grade, and lymph node positivity (p<0.0001 each, ). Subset analysis of ERG-negative and ERG-positive cancers revealed that these associations were driven by the subset of ERG-negative cancers and much less by ERG-positive cancers ( and ).
Figure 3 Association between CENPF staining and ERG expression by immunohistochemistry or TMPRSS2:ERG fusion by FISH.
Abbreviations: CENPF, centromere protein F; ERG, erythroblast transformation-specific (ETS)-related gene; TMPRSS2, transmembrane protease, serine 2; FISH, fluorescence in situ hybridization.
Association with other key genomic deletions
Earlier studies have provided evidence for distinct molecular subgroups of prostate cancers defined by TMPRSS2:ERG fusion and several genomic deletions. This includes a strong association of PTEN and 3p13 deletions with ERG positivity and of 5q21 and 6q15 deletions with ERG negativity.Citation23–Citation26 To study whether CENPF expression might be particularly associated with one of these genomic deletions, CENPF data were compared with preexisting findings on 10q23 (PTEN), 3p13 (FOXP1), 6q15 (MAP3K7), and 5q21 (CHD1) deletions. For tumors that were jointly analyzed, there were positive associations between CENPF overexpression and deletions of PTEN or 3p13 (p<0.0001 each), as well as negative associations between CENPF overexpression and deletions of 5q or 6q (p=0.027 and p<0.0001, respectively, ). These associations make intuitive sense due to the known link between these deletions and a positive ERG status (PTEN, 3p) or a negative ERG status (5q, 6q). However, subset analysis of ERG-negative and ERG-positive cancers demonstrated that CENPF overexpression was linked to PTEN deletions independent of the ERG status (p<0.0001 each, ).
Figure 4 Association between CENPF staining intensity and 10q23 (PTEN), 5q21 (CHD1), 6q15 (MAP3K7), and 3p13 (FOXP1) deletions in (A) all cancers, (B) the ERG-negative subset, and (C) the ERG-positive subset.
Abbreviations: CENPF, centromere protein F; PTEN, phosphatase and tensin homolog; CHD1, chromodomain-helicase-DNA-binding protein 1; MAP3K7, mitogen-activated protein kinase kinase kinase 7; FOXP1, fork head box protein P1.
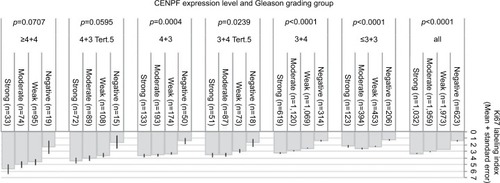
Association with tumor cell proliferation
High-level CENPF staining was linked to accelerated cell proliferation as measured by the Ki67-labeling index (Ki67LI) obtained from an earlier study.Citation27 The average Ki67LI increased from 1.41 in cancers lacking CENPF expression to 3.35 in cancers with strong CENPF levels (p<0.0001, ). This association was also found both in ERG-negative (p<0.0001) and ERG-positive cancers (p=0.0061; data not shown). It was independent from the Gleason score as it held true in all tumor subsets with identical Gleason score ().
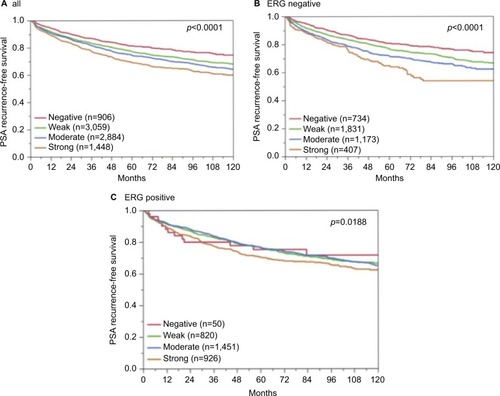
Association with PSA recurrence
Follow-up data were available for 8,298 patients with interpretable CENPF staining on the tissue microarray. A highly significant association between strong CENPF expression and early PSA recurrence was seen in all tumors (p<0.0001, ) and in the subset of ERG fusion-negative cancers (p<0.0001, ). The prognostic impact of CENPF was less significant in ERG fusion-positive cancers (p=0.0188, ). To better rate the prognostic power of CENPF, we performed further subset analyses in cancers with identical classical and quantitative Gleason scores. Here, CENPF staining did not provide clear-cut prognostic information beyond the Gleason score, neither in any subsets defined by the classical Gleason score (Figure S2A) nor in any subsets defined by the quantitative Gleason score (Figure S2B–H).
Figure 6 Association between CENPF expression and PSA recurrence after prostatectomy in (A) all cancers, (B) the ERG-negative subset, and (C) the ERG-positive subset. Abbreviations: CENPF, centromere protein F; PSA, prostate-specific antigen; ERG, erythroblast transformation-specific (ETS)-related gene.
Multivariate analyses
To evaluate whether CENPF expression can serve as a prognostic biomarker independent of the established prognostic factors (Gleason grade at biopsy, clinical stage, preoperative PSA level, Gleason grade at radical prostatectomy, pathological stage, surgical margin status, and nodal status), multivariate analyses of PSA recurrence-free survival were performed in 3 different scenarios (). CENPF proved to be a significant independent prognostic parameter in all 3 scenarios. The HR for PSA recurrence after prostatectomy peaked at 1.23 in the preoperative scenario 1. This held true for the ERG-negative subset with the exception of scenario 3 and the ERG-positive subset with the exception of scenario 2.
Table 2 HR for PSA recurrence-free survival after prostatectomy of established prognostic parameter and CENPF expression
Discussion
The results of our study identify CENPF as a strong and independent predictor of patient prognosis.
Successful analysis of >9,000 prostate cancers by IHC revealed detectable CENPF staining in 90% of prostate cancers, including almost 40% of tumors with no more than weak staining. The fact that such low-level staining was also found in normal prostate glands as well as in stromal cells suggests that our IHC protocol was sufficiently sensitive to detect physiological expression of CENPF. However, increased CENPF expression in 50% of cancers indicates that CENPF becomes upregulated during malignant transformation in a high fraction of prostate cancers. Only one published study has previously evaluated CENPF protein expression by IHC in prostate cancer. In line with our findings, Zhuo et al reported significantly higher CENPF levels in 99 tumor samples compared with benign tissues.Citation17 Tumor-associated upregulation of CENPF was also found in other tumor types including esophageal cancer, hepatocellular carcinoma, non-Hodgkin’s lymphoma, nasopharyngeal cancer, and breast cancer.Citation11–Citation17
CENPF overexpression was linked to features of aggressive and prognostically unfavorable cancers in our study, including advanced tumor stage, higher Gleason grade, presence of lymph node metastasis, accelerated cell proliferation, and early biochemical recurrence (p<0.0001 each). Our results are in line with earlier reports demonstrating an important role of CENPF for tumor cell proliferation in general and prostate cancer biology. For example, Laoukili et al identified CENPF as a direct target gene of the master cell cycle regulator FOXM1, which controls expression of many G2-specific genes and the activity of PI3K/AKT and MAPK signaling pathways.Citation28 More recently, cross-species regulatory network analysis described a synergistic interaction between FOXM1 and CENPF in driving prostate cancer malignancy and suggested that patients with overexpression of CENPF and FOXM1 had a particularly poor outcome.Citation10 High CENPF expression has also been linked to increased tumor aggressiveness and poor survival in esophageal squamous cell carcinoma as well as nasopharyngeal and breast cancer.Citation11,Citation12,Citation14
The molecular database attached to our tissue microarray allowed us to further study the impact of CENPF upregulation in molecularly defined subgroups of prostate cancers. About 50% of all prostate cancers carry a gene fusion linking the androgen-regulated serine protease TMPRSS2 with the ETS-transcription factor ERG resulting in an androgen-related overexpression of ERG.Citation21,Citation29,Citation30 We found a massive upregulation of CENPF in a particularly high fraction of ERG-positive cancers. The reasons underlying this effect are not known. The CENPF promoter does not carry an ERG binding site according to the eukaryotic promoter database,Citation31 but it cannot be excluded that CENPF is an indirect transcriptional target of ERG. For example, global expression analyses indicate upregulation of other members of the CENP family, including CENPO, CENPL, and CENPV in ERG-positive as compared to ERG-negative prostate cancers.Citation32 Further evidence comes from recent work identifying a regulatory network comprising miR-101, CoupTFII, FOXM1, and CENPF,Citation33 which has been suggested to interfere with ERG-dependent transcription.Citation34
Irrespective of the reasons leading to CENPF upregulation in ERG-positive cancers, our data demonstrate that the prognostic value of CENPF was reduced in ERG-positive cancers. In previous studies, using the same tissue microarray, we identified various proteins with higher expression levels in ERG-positive than in ERG-negative prostate cancers. With several of these, the prognostic impact was substantially stronger in ERG-negative than in ERG-positive cancers.Citation35–Citation37 The present study demonstrates that CENPF belongs to this type of protein. Other biomarkers were only prognostic in ERG-positive cancers.Citation38–Citation40 Overall, these data suggest that tumor-relevant functions of CENPF and other proteins can become modified in an ERG-positive molecular environment. This is conceivable as ERG activation leads to substantial changes of the intracellular environment, affecting the expression of >1,600 genes.Citation21,Citation29,Citation30
Next to TMPRSS2:ERG fusions, chromosomal deletions represent the second most frequent type of genomic aberration in prostate cancers, occurring at frequencies of up to 40%.Citation32,Citation41 In particular, deletions of PTEN (20%), 6q (20%), 5q (10%), and 3p (10%) belong to the most prevalent genomic alterations in this disease. These are linked to either positive (PTEN, 3p) or negative ERG status (6q, 5q) and are associated with poor patient prognosis.Citation23–Citation26 CENPF overexpression was unequivocally linked to deletions of PTEN only, suggesting a functional relationship between these two genes. PTEN deletion is the main cause for hyperactive PI3K/AKT signaling in prostate cancer and is associated with tumor growth, progression, and poor clinical outcome.Citation42 In fact, a relevant functional interaction between CENPF and PI3K/AKT signaling is supported by recent functional data demonstrating that PI3K/AKT signaling was completely abrogated when CENPF was silenced, together with FOXM1, in DU145 prostate cancer cells.Citation10
The results of our study identify CENPF as a promising candidate for a molecular test of prostate cancer aggressiveness. Our multivariate modeling suggests that a potential clinical application could be suitable to gain prognostic information not only at pretherapeutic stages, when only needle biopsies can be assessed, but also after radical prostatectomy when more comprehensive pathological data are available. It is of note that the Gleason grade is the strongest (and least expensive) prognostic feature in prostate cancer. In a recent analysis, we demonstrated that by using the percentage of unfavorable Gleason patterns, the Gleason grading could be transformed from a categorical into a continuous variable with an even finer discrimination of prognostic subgroups.Citation19,Citation43 The prognostic impact of CENPF expression largely disappeared in groups defined by Gleason grade categories or by comparable percentages of Gleason 4 patterns, demonstrating the power of morphological assessment of malignancy. These findings show that the threshold for a molecular test to significantly augment morphology in prognostication is rather high.
Limitations of the present study are that it was a retrospective study and that CENPF testing was done on prostatectomy specimens. To cope with the heterogeneity of prostate cancer, Kaplan–Meier analysis was done on 8,032 patients, which should guarantee a reliable result for the cohort. For individual decision-making, a prospective study is needed with at least 10 replicates of biopsy specimens for each proband.
Due to its critical role in cell proliferation and its frequent expression in cancer cells, CENPF gained interest as a potential molecular target for novel anticancer therapies. To date, no clinically applicable anti-CENPF drugs are available, but initial in vitro experiments are encouraging for the development of strategies for pharmacological inhibition. For example, zoledronic acid, a nitrogen-containing bisphosphonate usually applied in bone disease, is found to cause loss of CENPF from the kinetochore complex in breast cancer cell lines by inhibition of CENPF farnesylation/activation, subsequently leading to delayed cell cycle progression and inhibition of cell proliferation.Citation44 Moreover, due to the putative effect of CENPF on growth pathways, including PI3K/AKT and MAPK signaling,Citation10 CENPF inhibitors may provide an attractive means for inactivation of these signaling pathways.
Conclusion
In summary, our study shows the prognostic value of CENPF. The strong association between CENPF upregulation and PTEN deletion supports the concept of a regulatory function of CENPF for major growth pathways, which makes it an attractive candidate for novel therapeutic approaches.
Acknowledgments
The authors would like to thank W Fehrle for help in revision of the manuscript and are grateful to Christina Koop, Janett Lütgens, Sünje Seekamp, and Inge Brandt for excellent technical assistance. The Federal Ministry of Education and Research supported this work (grant no. 01KU1505B). An abstract of the present data was published by Lebok et al in Pathologe.Citation45
Supplementary materials
Figure S1 Prognostic impact of ERG expression on biochemical relapse.
Abbreviation: ERG, erythroblast transformation-specific (ETS)-related gene.
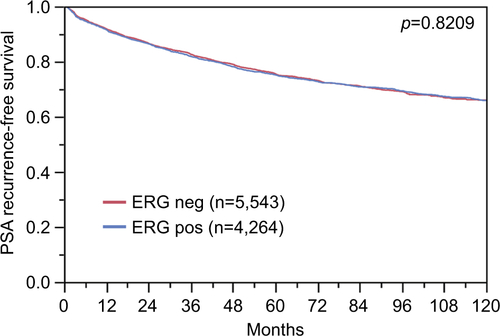
Figure S2 Prognostic impact of CENPF expression in subsets of cancers defined by the Gleason score.
Notes: (A) Impact of negative and positive CENPF expression compared with the classical Gleason score categories. (B–H) Impact of negative and positive CENPF expression compared with the quantitative Gleason score categories defined by subsets of cancers with (B) ≤5% Gleason 4 patterns, (C) 6%–10% Gleason 4 patterns, (D) 11%–20% Gleason 4 patterns, (E) 21%–30% Gleason 4 patterns, (F) 31%–49% Gleason 4 patterns, (G) 50%–60% Gleason 4 patterns, (H) 61%–100% Gleason 4 patterns.
Abbreviation: CENPF, centromere protein F.
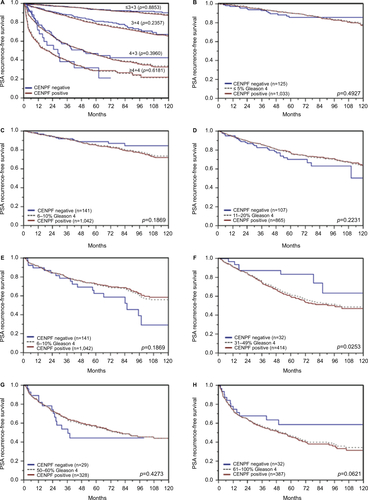
Table S1 Association between CENPF staining results and prostate cancer phenotype in ERG-negative tumors
Table S2 Association between CENPF staining results and prostate cancer phenotype in ERG-positive tumors
Author contributions
CS, AML, CHM, RS, and GS designed the study and drafted the manuscript. TS, HH, MG, and CÖ participated in study design. AH, EN, and FJ performed IHC analysis and scoring. MK, UM, DP, and RS participated in pathology data analysis. CS and RS performed statistical analysis. GS, CMK, and MK participated in data interpretation and helped to draft the manuscript. All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- TorreLABrayFSiegelRLFerlayJLortet-TieulentJJemalAGlobal cancer statistics, 2012CA Cancer J Clin20156528710825651787
- WiltTJBrawerMKJonesKMProstate Cancer Intervention versus Observation Trial (PIVOT) Study GroupRadical prostatectomy versus observation for localized prostate cancerN Engl J Med2012367320321322808955
- ThompsonIMJrTangenCMProstate cancer: uncertainty and a way forwardN Engl J Med2012367327027122808963
- ZhaoMa LZhuXXMitosin/CENP-F in mitosis, transcriptional control, and differentiationJ Biomed Sci200613220521316456711
- LiaoHWinkfeinRJMackGRattnerJBYenTJCENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosisJ Cell Biol199513035075187542657
- ZhuXManciniMAChangKHCharacterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progressionMol Cell Biol1995159501750297651420
- JohnsonVLScottMIHoltSVHusseinDTaylorSSBub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congressionJ Cell Sci2004117(Pt 81577158915020684
- PooleyRDMoynihanKLSoukoulisVMurine CENPF interacts with syntaxin 4 in the regulation of vesicular transportJ Cell Sci2008121Pt 203413342118827011
- PooleyRDReddySSoukoulisVRolandJTGoldenringJRBaderDMCytLEK1 is a regulator of plasma membrane recycling through its interaction with SNAP-25Mol Biol Cell20061773176318616672379
- AytesAMitrofanovaALefebvreCCross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancyCancer Cell201425563865124823640
- O’BrienSLFaganAFoxEJCENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancerInt J Cancer200712071434144317205517
- CaoJYLiuLChenSPPrognostic significance and therapeutic implications of centromere protein F expression in human nasopharyngeal carcinomaMol Cancer2010923720828406
- DaiYLiuLZengTCharacterization of the oncogenic function of centromere protein F in hepatocellular carcinomaBiochem Biophys Res Commun2013436471171823791740
- MiYJGaoJXieJDPrognostic relevance and therapeutic implications of centromere protein F expression in patients with esophageal squamous cell carcinomaDis Esophagus201326663664323163484
- KoonNSchneider-StockRSarlomo-RikalaMMolecular targets for tumour progression in gastrointestinal stromal tumoursGut200453223524014724156
- ErlansonMCasianoCATanEMLindhJRoosGLandbergGImmunohistochemical analysis of the proliferation associated nuclear antigen CENP-F in non-Hodgkin’s lymphomaMod Pathol199912169749950165
- ZhuoYJXiMWanYPEnhanced expression of centromere protein F predicts clinical progression and prognosis in patients with prostate cancerInt J Mol Med201535496697225647485
- SchlommTIwersLKirsteinPClinical significance of p53 alterations in surgically treated prostate cancersMod Pathol200821111371137818552821
- SauterGSteurerSClauditzTSClinical utility of quantitative Gleason grading in prostate biopsies and prostatectomy specimensEur Urol201669459259826542947
- KononenJBubendorfLKallioniemiATissue microarrays for high-throughput molecular profiling of tumor specimensNat Med1998478448479662379
- WeischenfeldtJSimonRFeuerbachLIntegrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancerCancer Cell201323215917023410972
- MinnerSEnodienMSirmaHERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of anti-hormonal therapyClin Cancer Res201117185878588821791629
- BurkhardtLFuchsSKrohnACHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancerCancer Res20137392795280523492366
- KluthMHesseJHeinlAGenomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusionsMod Pathol201326797598323370768
- KrohnADiedlerTBurkhardtLGenomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancerAm J Pathol2012181240141222705054
- KrohnASeidelABurkhardtLRecurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancersJ Pathol2013231113014123794398
- TennstedtPKösterPBrüchmannAThe impact of the number of cores on tissue microarray studies investigating prostate cancer biomarkersInt J Oncol201240126126821956230
- LaoukiliJKooistraMRBrásAFoxM1 is required for execution of the mitotic programme and chromosome stabilityNat Cell Biol20057212613615654331
- TomlinsSARhodesDRPernerSRecurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancerScience2005310574864464816254181
- BraseJCJohannesMMannspergerHTMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-β signalingBMC Cancer20111150722142399
- DreosRAmbrosiniGPérierRCBucherPThe eukaryotic promoter database: expansion of EPDnew and new promoter analysis toolsNucleic Acids Res201543Database issueD92D9625378343
- TaylorBSSchultzNHieronymusHIntegrative genomic profiling of human prostate cancerCancer Cell2010181112220579941
- LinSCKaoCYLeeHJDysregulation of miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of prostate cancerNat Commun201671141827108958
- Vlaeminck-GuillemVVanackerJMVergerAMutual repression of transcriptional activation between the ETS-related factor ERG and estrogen receptorOncogene200322508072808414603248
- GruppKJedrzejewskaKTsourlakisMCHigh mitochondria content is associated with prostate cancer disease progressionMol Cancer201312114524261794
- GruppKOspina-KlinckDTsourlakisMCNY-ESO-1 expression is tightly linked to TMPRSS2-ERG fusion in prostate cancerProstate201474101012102224789172
- StummLBurkhardtLSteurerSStrong expression of the neuronal transcription factor FOXP2 is linked to an increased risk of early PSA recurrence in ERG fusion-negative cancersJ Clin Pathol201366756356823559350
- BurdelskiCBujupiETsourlakisMCLoss of SOX9 expression is associated with PSA recurrence in ERG-positive and PTEN deleted prostate cancersPLoS One2015106e012852526030748
- BurdelskiCMenanDTsourlakisMCThe prognostic value of SUMO1/Sentrin specific peptidase 1 (SENP1) in prostate cancer is limited to ERG-fusion positive tumors lacking PTEN deletionBMC Cancer20151553826202067
- GruppKBoumesliRTsourlakisMCThe prognostic impact of high Nijmegen breakage syndrome (NBS1) gene expression in ERG-negative prostate cancers lacking PTEN deletion is driven by KPNA2 expressionInt J Cancer201413561399140724510842
- SunJLiuWAdamsTSDNA copy number alterations in prostate cancers: a combined analysis of published CGH studiesProstate200767769270017342750
- WangYHeXNgeowJEngCGATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancerHum Mol Genet201221356957622021428
- SauterGClauditzTSteurerSIntegrating tertiary Gleason 5 patterns into quantitative Gleason grading in prostate biopsies and prostatectomy specimensEur Urol201873567468328117112
- BrownHKOttewellPDColemanREHolenIThe kinetochore protein Cenp-F is a potential novel target for zoledronic acid in breast cancer cellsJ Cell Mol Med201115350151320015195
- LebokPÖzdenCSchlommTUpregulation of centromer protein F (CENPF) is linked to an aggressive subset of ERG-negative prostate cancersPathologe2017385482