Abstract
Purpose
DNMT3A and NPM1 mutations are known to impact the prognosis of acute myeloid leukemia (AML). DNMT3A mutations are negative prognostic factors, while NPM1 mutations are low-risk factors and inclined to concurrently appear with DNMT3A mutations. In this study, we aimed to find out how NPM1 mutations affect patients’ outcomes in the background of DNMT3A mutations.
Patients and methods
We screened The Cancer Genome Atlas (TCGA) database and found 51 AML patients with DNMT3A mutations. Of them, 28 patients had a combination of NPM1 mutations.
Results
In all, NPM1 had the highest mutation frequency (n=28, 54.9%). DNMT3Amut/NPM1mut patients had higher bone marrow (BM) blasts (P=0.015), higher FLT3-ITD/TKD rate (P=0.004), and lower IDH2 mutation rate (P=0.014) than the DNMT3Amut/NPM1wild patients, while their prognoses were the same as the DNMT3Amut/NPM1wild patients (P>0.1). All 51 patients benefited from hematopoietic stem cell transplantation (HSCT) treatment (P=0.005 and 0.001 for event-free survival [EFS] and overall survival [OS], respectively). In the 23 patients with DNMT3Amut/NPM1wild, those who received HSCT had prolonged EFS and OS (P=0.043 and 0.008, respectively), while HSCT treatment did not produce a positive impact on EFS and OS in the remaining 28 patients with DNMT3Amut/NPM1mut (P=0.056 and 0.053, respectively).
Conclusion
Our study found that NPM1 mutations influenced BM blasts’ percentage, FLT3-ITD/TKD rate, and IDH2 mutation rate in AML patients with DNMT3A mutations but made little difference to the overall prognosis. While HSCT treatments benefited all DNMT3Amut patients, it was better for DNMT3Amut/NPM1wild patients to extend their EFS and OS.
Introduction
Acute myeloid leukemia (AML) is a kind of heterogeneous disease characterized by the clonal expansion of hematopoietic stem cells (HSCs) or hematopoietic progenitor cells (HPCs) in the bone marrow (BM), blood, and other tissues without differentiation.Citation1
As sequencing methods have technologically advanced in the past four decades, next-generation sequencing (NGS) is now available for a detailed understanding of the molecular pathogenesis of AML.
DNMTs are genes that encode DNMTs for the methylation of CpG islands. As methylations reduce the expression of downstream genes, spontaneous defects in DNMTs lead to the instability of genome structure and thus increase the possibility of cancer.Citation2 Since Ley et alCitation3 confirmed that DNMT3A mutations were highly recurrent in patients with de novo AML and associated with a poor outcome, many prognostic studies about DNMT3A-mutated AML have been carried out.Citation4–Citation10 Occurring in ~20% of AML patients and having a high proportion of R882 mutation,Citation11,Citation12 DNMT3A mutations are often accompanied by FLT3-ITD, NPM1, IDH1, and IDH2 mutations.Citation13 NPM1 mutations were more common in patients with DNMT3A mutations than in DNMT3A wild-type patients.Citation14 High-risk NPM1/FLT3 mutation (NPM1wild/FLT3-ITDnegative, NPM1wild/FLT3-ITDpositive, or NPM1mut/FLT3-ITDpositive) was thought to be related with poor prognosis.Citation5 Hematopoietic stem cell transplantation (HSCT) has been shown to benefit the survival of all AML patients with DNMT3A mutations.Citation8,Citation9 Gale et alCitation15 argued that the presence of DNMT3A mutations should be considered as a poor-risk prognostic factor, irrespective of the NPM1 genotype as it may disturb the outcome. There have been few studies concerning DNMT3A mutations alone in AML patients together with studying the biological and clinical features with companion genes. Therefore, understanding the biological and clinical characteristics of these patients with DNMT3A mutations is important for estimating their event-free survival (EFS) and overall survival (OS) and likely advantageous in deciding the most suited clinical treatment.
Patients and methods
Patients
We selected and enrolled 51 patients with diagnosed DNMT3A-mutated AML from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). We collected data including age, gender, AML French-American-British classification subtypes, karyotype, cytogenetics’ risk, white blood cell (WBC) count, BM blasts percentage, peripheral blood (PB) blasts percentage, relapse, mutated recurrent genes, and combined mutation genes, which were highly related to AML such as FLT3, IDH1/2, TET2, NRAS, CEBPA, PTPN11, KRAS, U2AF1, SMC1A, SMC3, and RAD21, with their clinical figures to include EFS and OS. Gene mutations were detected by NGS. Written informed consent was obtained from all patients. This study was approved by the Human Research Ethics Committee of Washington University.
Statistical analyses
The end points were EFS and OS. EFS is the time from the date of diagnosis to removal from the study due to the absence of complete remission, relapse, or death. OS is the time from the date of diagnosis to death due to any cause.
We compared different biological and clinical characteristics by using different statistical methods. The Student’s t-test was applied to two group comparisons, and Chi-square test was used to compare the rate between them. Survival analysis was performed by Kaplan–Meier method. A two-sided P-value of <0.05 was considered statistically significant. All statistical analyses were performed by the SPSS Version 20.0 software.
Results
Biological and clinical characteristics
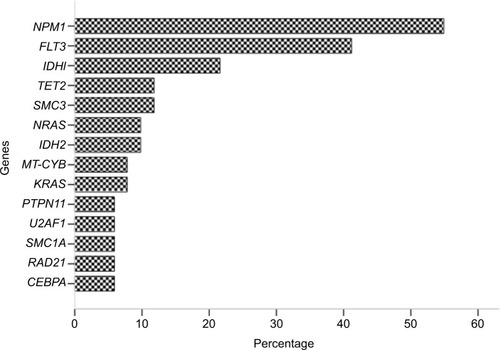
In all of the 51 patients, NPM1 was the most frequently combined mutation gene (n=28, 54.9%), followed by FLT3 (n=21, 41.2%), IDH1 (n=11, 21.6%), and TET2 (n=6, 11.8%). The mutational spectrum of all genes with >5% mutation frequency is shown in . The biological and clinical characteristics are summarized in .
Table 1 Biological and clinical characteristics
Figure 1 Mutational spectrum of all genes with >5% mutation frequency.
Note: NPM1 was the most frequently combined mutation gene (n=28, 54.9%), followed by FLT3 (n=21, 41.2%), IDH1 (n=11, 21.6%), TET2 (n=6, 11.8%), SMC3 (n=6, 11.8%), IDH2 (n=5, 9.8%), NRAS (n=5, 9.8%), KRAS (n=4, 7.8%), MT-CYB (n=4, 7.8%), CEBPA (n=3, 5.9%), PTPN11 (n=3, 5.9%), U2AF1 (n=3, 5.9%), SMC1A (n=3, 5.9%), and RAD21 (n=3, 5.9%).
Patients were divided into two groups depending on their NPM1 mutation states. There were 23 patients with DNMT3Amut/NPM1wild and 28 patients with DNMT3Amut/NPM1mut. The median age of the cohort was 58 years (24 men and 27 women; age range: 21–81 years), and 47.1% of the patients were ≥60 years. WBC counts ranged from 1.2 to 298.4×109/L with a median of 45.0×109/L, and 19 cases were not <50×109/L. The median BM blast percentage and PB blast percentage were 76 and 36%, respectively, and ranged from 32 to 100% and 0 to 97%; there were 30 (58.8%) cases with BM blasts ≥70% and six (30.0%) cases with PB blasts ≥70%. In 28 of the 51 patients (58.8%) who had relapses, days from collection to first relapse ranged from 53 to 1230 days with a median of 324.7 days. There were two (8.7%) cases with FLT3-ITD mutation and two (8.7%) cases with FLT-TKD mutation. HSCT treatments were received by 23 (45.1%) cases.
Upon comparing the two groups with DNMT3A mutations, we found that patients with DNMT3Amut/NPM1mut had higher BM blasts (P=0.015), higher FLT3-ITD/TKD (P=0.004), and lower IDH2 mutation (P=0.014). Other biological or clinical characteristics showed no significant differences between the two groups.
Comparison of EFS and OS between different biological and clinical characteristics
To determine what kind of biological or clinical characteristics were easily influenced by NPM1 mutation, we chose the following parameters for EFS and OS analyses: age (<60 vs ≥60 years), cytogenetic risk (intermediate vs poor), WBC count (<50×109 vs ≥50×109/L), BM blasts percentage (<70 vs ≥70%), PB blasts percentage (<70 vs ≥70%), mutated recurrent genes (<5 vs ≥5), genes with no <10 (19.6%) cases harboring FLT3-ITD/TKD and IDH1 changes (mutation vs wild type), and HSCT (yes vs no). The results are shown in .
Table 2 Kaplan–Meier analysis for EFS and OS between different biological and clinical character groups
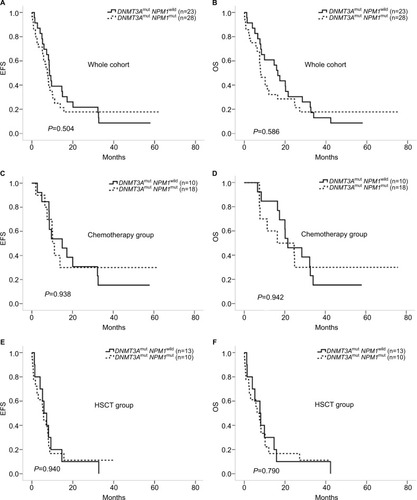
The NPM1 mutation had no influence on any of the abovementioned parameters in DNMT3A-mutated AML patients. EFS and OS between DNMT3Amut/NPM1wild and DNMT3Amut/NPM1mut groups showed no difference (P=0.504 and 0.586, respectively, ). Furthermore, there was also no difference in EFS and OS between groups of patients with only chemical therapy (P=0.938 for EFS and P=0.942 for OS, ) or HSCT treatment (P=0.940 for EFS and P=0.790 for OS, ).
Figure 2 Comparison of EFS and OS between different biological and clinical character groups.
Notes: (A and B) EFS and OS of DNMT3Amut/NPM1wild vs DNMT3Amut/NPM1mut in all of the 51 patients. (C and D) EFS and OS of DNMT3Amut/NPM1wild vs DNMT3Amut/NPM1mut in 28 patients with chemotherapy. (E and F) EFS and OS of DNMT3Amut/NPM1wild vs DNMT3Amut/NPM1mut in 23 patients with HSCT.
Abbreviations: EFS, event-free survival; HSCT, hematopoietic stem cell transplantation; OS, overall survival.
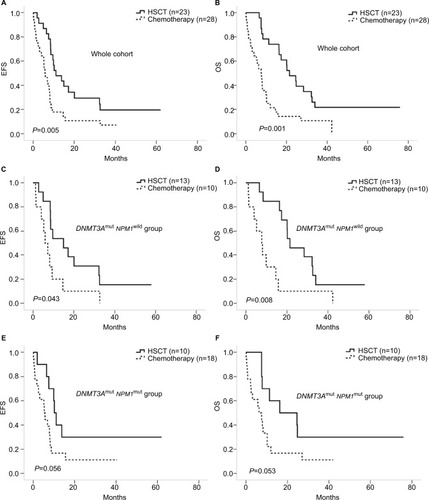
HSCT was an effective way for the treatment of DNMT3A-mutated AML patients (P=0.005 for EFS and P=0.001 for OS) in all of the 51 patients (). We also compared the influence of different treatments in each group. The group of people with DNMT3Amut/NPM1wild derived the best of HSCT treatment and had longer EFS and OS than patients who received only chemical therapy (P=0.043 for EFS and P=0.008 for OS, ). Patients in the other group with DNMT3Amut/NPM1mut did not show similar results (P=0.056 for EFS and P=0.053 for OS, ).
Figure 3 Comparison of chemotherapy and HSCT in different mutation groups.
Notes: (A and B) EFS and OS of all the 51 patients. (C and D) EFS and OS of the 23 patients with DNMT3Amut/NPM1wild. (E and F) EFS of the 28 patients with DNMT3Amut/NPM1mut.
Abbreviations: EFS, event-free survival; HSCT, hematopoietic stem cell transplantation; OS, overall survival.
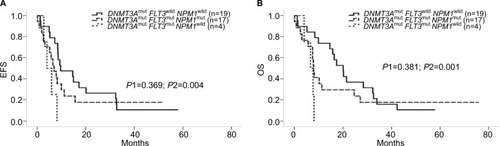
For further detailed analysis, we considered another subgroup of patients with DNMT3Amut/FLT3mut/NPM1wild from the existing cohort of 51 patients. We compared the EFS and OS between DNMT3Amut/FLT3wild/NPM1wild patients (n=19) and DNMT3Amut/FLT3mut/NPM1wild patients (n=4) and found that there were significant differences between these two groups (P2=0.004 for EFS in and P2=0.001 for OS in ), but there was no difference between the outcomes for the DNMT3Amut/FLT3wild/NPM1wild and DNMT3Amut/FLT3mut/NPM1mut groups (n=17, P1>0.1, ).
Figure 4 Influences of FLT3-ITD/TKD to the cohort.
Notes: (A and B) EFS and OS of DNMT3Amut/FLT3wild/NPM1wild vs DNMT3Amut/FLT3mut/NPM1mut vs DNMT3Amut/FLT3mut/NPM1wild; P1 for comparison between DNMT3Amut/FLT3wild/NPM1wild and DNMT3Amut/FLT3mut/NPM1mut; P2 for comparison between DNMT3Amut/FLT3wild/NPM1wild vs DNMT3Amut/FLT3mut/NPM1wild.
Abbreviations: EFS, event-free survival; OS, overall survival.
Discussion
Our study showed that DNMT3A mutations in AML had higher BM blasts, higher FLT3-ITD/TKD mutation rate, and higher IDH2 mutation rate when combined with NPM1 mutation, but little difference in prognoses compared with NPM1 wild type. HSCT treatment benefited all patients who carried the DNMT3A mutation and was a better choice for the DNMT3Amut/NPM1wild group.
In Gale et al’sCitation15 study, 80% of 272 patients with DNMT3A mutation concomitantly had the NPM1 mutation. We analyzed the biological and clinical features of 51 patients with DNMT3A mutations and found NPM1 to be the most common accompanying mutation (n=28, 54.9%). This may be due to a relatively high percentage of patients older than 60 years in our study (47.1 vs 3%). Many genes participate in AML development.Citation16 Mutations in NPM1, IDH2, and biallelic CEBPA are favorable risk factors;Citation13 while FLT3-ITD positive, IDH1, TET2,Citation13 KRAS,Citation17 U2AF1, and PTPN11 are always associated with poor outcomes.Citation18,Citation19 Although showing different mutation rates in AML patients, genes such as NRAS,Citation20 SMC1A, SMC3, and RAD21 showed no influence in the prognosis.Citation21
Holmberg et alCitation22 did a basic clinical experiment on the relationship between DNMT3A and NPM1 and found that deficiency in DNMT3A enhanced the rearrangement of heterochromatin, which was triggered by the loss of NPM1. In our study, 35.7% of DNMT3Amut/NPM1mut patients harbored the FLT3-ITD mutation and this rate was similar to that reported by Gale et alCitation15 (37%). Thol et alCitation5 also found that patients with DNMT3A mutations were more likely to have mutations in NPM1 with a trend toward a higher FLT3 mutation rate. In our study, the IDH2 mutation was seen in 21.7% patients in the DNMT3Amut/NPM1wild group, while those in the DNMT3Amut/NPM1mut group did not harbor the IDH2 mutation. Another study showed that the percentage of IDH2 mutations were 11.5 and 33.3% for DNMT3Amut/NPM1wild and DNMT3Amut/NPM1mut, respectively.Citation15 We were not very consistent with their IDH2 mutation rates, and this may be related to the small number of cases.
Comparison of EFS and OS between different biological and clinical characteristics groups showed no difference. Similar to Xu et al’sCitation23 conclusion, we also confirmed that HSCT is a better option for patients with DNMT3A mutations. We found that chemical therapy and HSCT treatment had the same effect in the DNMT3Amut/NPM1mut group. We supposed that this may be due to high mutation rate of FLT3 in the NPM1-mutated group. Previous studies suggested that FLT3-ITD always results in significantly worse clinical outcomes and weakens the curative effect of conventional chemotherapy,Citation24 as well as shortening the EFS and OS in DNMT3A-mutated patients.Citation25 Only the R140 mutation in IDH2 seems to have prognostic meaning and is associated with a better outcome.Citation13 In our study, we found that IDH2 mutation only occurred in DNMT3Amut/NPM1wild patients and three of the five IDH2 mutations happened at the R140 locus. This could be another reason why the DNMT3Amut/NPM1mut group showed similar results to the DNMT3Amut/NPM1wild group.
There are some limitations to our study. First, the small sample size may have reduced the accuracy of our results. Second, our study has a retrospective design, the effectiveness of which is limited when compared to a prospective study.
Conclusion
All patients with the DNMT3A mutation benefited from HSCT, but those who also have NPM1 mutations should be treated carefully given their high FLT3-ITD/TKD rate, and hence, HSCT may not be better than chemotherapy for this group of patients.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81500118 and 61501519), the China Postdoctoral Science Foundation funded project (project no 2016M600443), and PLAGH project of Medical Big Data (project no 2016MBD-025).
Disclosure
The authors report no conflicts of interest in this work.
References
- DohnerHWeisdorfDJBloomfieldCDAcute myeloid leukemiaN Engl J Med2015373121136115226376137
- EstellerMEpigenetics in cancerN Engl J Med2008358111148115918337604
- LeyTJDingLWalterMJDNMT3A mutations in acute myeloid leukemiaN Engl J Med2010363252424243321067377
- LinNFuWZhaoCLiBYanXLiYBiologico-clinical significance of DNMT3A variants expression in acute myeloid leukemiaBiochem Biophys Res Commun20174941–227027729024628
- TholFDammFLudekingAIncidence and prognostic influence of DNMT3A mutations in acute myeloid leukemiaJ Clin Oncol201129212889289621670448
- GaidzikVIWeberDPaschkaPGerman-Austrian Acute Myeloid Leukemia Study Group (AMLSG)DNMT3A mutant transcript levels persist in remission and do not predict outcome in patients with acute myeloid leukemiaLeukemia2017321303728643785
- SunYShenHXuTPersistent DNMT3A mutation burden in DNMT3A mutated adult cytogenetically normal acute myeloid leukemia patients in long-term remissionLeuk Res20164910210727626217
- TieRZhangTFuHAssociation between DNMT3A mutations and prognosis of adults with de novo acute myeloid leukemia: a systematic review and meta-analysisPLoS One201496e9335324936645
- ShivarovVGueorguievaRStoimenovATiuRDNMT3A mutation is a poor prognosis biomarker in AML: results of a meta-analysis of 4500 AML patientsLeuk Res201337111445145023962568
- LiYZhuBAcute myeloid leukemia with DNMT3A mutationsLeuk Lymphoma20145592002201224283755
- YuanXQPengLZengWJJiangBYLiGCChenXPDNMT3A R882 mutations predict a poor prognosis in AML: a meta-analysis from 4474 patientsMedicine (Baltimore)20169518e351927149454
- BerensteinRBlauIWSuckertNQuantitative detection of DNMT3A R882H mutation in acute myeloid leukemiaJ Exp Clin Cancer Res2015345525994761
- YoheSMolecular genetic markers in acute myeloid leukemiaJ Clin Med20154346047826239249
- ElGDTaalabMMGhazyHFEneenAFDNMT3A R882 mutations in patients with cytogenetically normal acute myeloid leukemia and myelodysplastic syndromeBlood Cells Mol Dis2014531–2616624512939
- GaleRELambKAllenCSimpson’s paradox and the impact of different DNMT3A mutations on outcome in younger adults with acute myeloid leukemiaJ Clin Oncol201533182072208325964253
- LeyTJMillerCDingLCancer Genome Atlas Research NetworkGenomic and epigenomic landscapes of adult de novo acute myeloid leukemiaN Engl J Med2013368222059207423634996
- ZhouJDYaoDMLiXXKRAS overexpression independent of RAS mutations confers an adverse prognosis in cytogenetically normal acute myeloid leukemiaOncotarget2017839660876609729029494
- OhgamiRSMaLMerkerJDNext-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutationsMod Pathol201528570671425412851
- AndradeFGNoronhaEPBrissonGDBrazilian Study Group of Childhood Acute Myeloid Leukemia (IMol-AMLBSG) as coauthorsMolecular characterization of pediatric acute myeloid leukemia: results of a multicentric study in BrazilArch Med Res201647865666728476193
- BacherUHaferlachTSchochCKernWSchnittgerSImplications of NRAS mutations in AML: a study of 2502 patientsBlood2006107103847385316434492
- TholFBollinRGehlhaarMMutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implicationsBlood2014123691492024335498
- HolmbergOKNisterMLindstromMSLoss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcriptionJ Biol Chem201428950346013461925349213
- XuYSunYShenHAllogeneic hematopoietic stem cell transplantation could improve survival of cytogenetically normal adult acute myeloid leukemia patients with DNMT3A mutationsAm J Hematol2015901199299726223865
- FrohlingSSchlenkRFBreitruckJAML Study Group UlmAcute myeloid leukemia. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group UlmBlood2002100134372438012393388
- TangSShenHMaoXFLT3-ITD with DNMT3A R882 double mutation is a poor prognostic factor in Chinese patients with acute myeloid leukemia after chemotherapy or allogeneic hematopoietic stem cell transplantationInt J Hematol2017106455256128616699
