Abstract
Background
Lung cancer is the leading cause of cancer-related death worldwide and it is critical to discover specific biomarkers to provide better individualized treatment and subsequently better prognosis. The sirtuins (SIRT1–7) have been reported to be involved in cancers including non-small cell lung cancer (NCSLC), however, the results are not consistent and not all the seven sirtuins are explored and compared.
Methods
TCGA data was downloaded and used to investigate and compare the associations of sirtuins mRNA levels with clinicopathological parameters and prognosis in NSCLC.
Results
Our results suggested SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in adeno-carcinoma (ADC) patients and female patients while SIRT5 were highly expressed in squamous cell carcinoma (SCC) patients and male patients. Associations of high SIRT7 with younger onset age, high SIRT1 with distant metastasis and low T stage, and high SIRT4 with high T stage and TNM stage were also found. Kaplan-Meier plot curves and univariate Cox proportional regression analyses indicated that high SIRT2, SIRT4, and SIRT6 expressions were associated with longer overall survival (OS) time. Multivariate analyses indicated that SIRT2 and SIRT6 were still associated with OS. For recurrence-free survival (RFS), high SIRT1 expression was significantly associated with shorter RFS time while high SIRT2-3 and SIRT5-7 expressions were associated with longer RFS time in univariate analyses. After adjusting the confounding factors, significant associations were still found in SIRT1-2 and SIRT5-7 but not in SIRT3. We also stratified the patients by combining SIRT1 and SIRT2 and revealed that the combination of SIRT1 and SIRT2 was a better prediction model for RFS in NSCLC. To preliminarily understand the potential mechanisms of sirtuins in NSCLC carcinogenesis, the genes co-expressed with sirtuins were analyzed and annotated.
Conclusion
sirtuins might be the potential therapy targets and prognostic biomarkers in NSCLC.
Introduction
Lung cancer is the leading cause of cancer-related death worldwide with a low 5-year survival rate of 18%.Citation1 Small cell lung cancer (SCLC) and non–small cell lung cancer (NSCLC) are the two major subtypes of lung cancer.Citation2 NSCLC accounts for ~85% of all lung cancers.Citation2 Surgical resection is an effective approach to achieve long-term survival; however, the prognosis of NSCLC remains poor due to late diagnosis and postoperative relapse.Citation3 Thus, it is critical to discover specific biomarkers to provide better individualized treatment and better prognosis.
In the past decades, a larger number of cellular pathways such as the EGFR signaling pathway have been identified in the carcinogenesis of a series of malignancies including lung cancer and are used as targets in drug development and predictive factors of prognosis.Citation4,Citation5 This encourages researchers to continue to identify novel therapy targets and prognostic biomarkers.
The sirtuins are a family of mammalian proteins that are homologous to the yeast silent information regulator 2 (Sir2).Citation6,Citation7 Seven different sirtuins (SIRT1–7) have been found to be widely expressed in mammals and exert nicotinamide adenine dinucleotide (NAD+)-dependent protein deacylase and/or ADP ribosyltransferase activities.Citation8–Citation16 In addition to their deacylase activities, recent studies have suggested SIRT5 can mediate protein desuccinylation, demalonylation, and deglutarylation,Citation17–Citation20 and SIRT7 can mediate histone desuccinylation.Citation21 The sirtuins have been linked to the modulation of numerous critical biological processes including cell metabolism, genomic stability, cell cycle, cell division, differentiation, and transcriptional regulation and are also correlated with the pathogenesis of a variety of diseases such as metabolic diseases,Citation22 neurode-generative diseases,Citation23 aging,Citation24 and cardiovascular diseases.Citation25 Currently, accumulating evidence has shown that the aberrant epigenetic activation of sirtuin signaling pathways contributes to tumor carcinogenesis and may be potential therapeutic targets for future treatments and biomarkers in predicting prognosis in cancers.Citation26,Citation27 Furthermore, some previous studies have been conducted to explore the clinical significance of sirtuins in lung cancer. However, the results are not consistent, the sample sizes in the previous studies are limited, and not all of the seven sirtuins were explored and compared.Citation28–Citation42
In the present study, we utilized the mRNA profile dataset of 1017 NSCLC patients from The Cancer Genome Atlas (TCGA) to investigate and compare the associations of the seven sirtuins (SIRT1–7) with clinicopathological parameters and prognosis in NSCLC and preliminarily explore the molecular mechanisms of sirtuins in carcinogenesis.
Materials and methods
Analysis of TCGA data
To investigate the associations of sirtuin mRNA levels with clinicopathological parameters and clinical outcomes in NSCLC, we downloaded NSCLC mRNA profile data and corresponding clinical data from the TCGA database. The patient samples were divided into two groups by the optimal cutoff value of sirtuin mRNA levels that was obtained on the basis of prognosis. The correlations between sirtuin expression and clinicopathological parameters were analyzed by the chi-square or Fisher’s exact test. Kaplan–Meier plot curves and Cox proportional regression analysis were used to analyze the effects of sirtuins on overall survival (OS) and recurrence-free survival (OS) in NSCLC. Statistical analyses were conducted with the software GraphPad Prism 6 and SPSS 19.0. P-value <0.05 was considered to be statistically significant. To investigate the related genes during sirtuin alteration that might be involved in lung cancer carcinogenesis, the expression correlations of sirtuins with other genes were examined by Pearson’s correlation analyses. Then, the top 500 genes that correlated with sirtuins were subjected to gene ontology (GO) analysis in the GeneCoDis3 online database (http://genecodis.cnb.csic.es).
Literature review
A systematic literature search was conducted in three English databases (PubMed, EMBASE, and ISI Web of science) for related reports on the associations of sirtuin expression and OS in NSCLC up to January 2018. The following terms were used: “lung OR pulmonary”, “cancer OR tumor OR carcinoma”, and “Sirtuins OR SIRT1 OR SIRT2 OR SIRT3 OR SIRT4 OR SIRT5 OR SIRT6 OR SIRT7”. The retrieved documents were screened by two independent authors by reviewing the article titles, abstracts, and full texts. In addition, the review articles were screened to identify additional eligible studies. The sample size, sirtuin detection method, and the HRs with 95% CIs of each included study were extracted by two investigators independently.
Examination of the concordance of SIRT2 mRNA and protein expressions in NSCLC
To investigate the concordance of SIRT2 mRNA and protein expressions in NSCLC, the SIRT2 protein and mRNA expressions in NSCLC and normal lung tissues were initially compared. Results of SIRT2 immunohistochemical (IHC) staining of NSCLC and normal tissues were evaluated using the data from the Human Protein Atlas (HPA).Citation43 SIRT2 mRNA expression in NSCLC and normal adjacent tissues was compared with the TCGA data by the Mann–Whitney test. In addition, we undertook a re-analysis of the results from Li et al’s study, which detected SIRT2 mRNA expressions by quantitative real-time polymerase chain reaction (qRT-PCR) and protein expressions by Western blotting in seven NSCLC tissues and corresponding normal tissues.Citation44 The mRNA data were obtained by reading the histograms with Plot Digitizer software. In addition, Image J 1.8.0 software was used to scan the Western blotting bands to obtain expression levels of SIRT2 protein. Then, SIRT2 mRNA and protein expressions in NSCLC and corresponding normal tissues were compared with the paired t-test. Next, the correlations of SIRT2 mRNA with protein expressions in Li et al’ report were explored by Pearson’s correlation analysis. At the same time, studies reporting both proteomics and transcription profiles in the same lung cancer patients were screened by searching the PRIDE database and reading the corresponding publications. Two studies were identified.Citation45,Citation46 However, only one study reported SIRT2.Citation46 Then, SIRT2 mRNA and protein expression data were extracted and subjected to Pearson’s correlation analysis. P<0.05 was considered as indicative of statistical significance.
Results
Correlations of sirtuins with clinicopathological parameters in NSCLC
In order to explore the associations of sirtuin mRNA expressions with the clinicopathological parameters of NSCLC patients, we downloaded the related datasets from the TCGA database. In total, 1017 NSCLC patients with basal clinical characteristics and prognosis information were identified. The sirtuin expressions of the patients were split into the high-expression and low-expression group according to the optimal cutoff value that was obtained on the basis of prognosis. The chi-square test was used to clarify the associations of sirtuins with the clinicopathological parameter of NSCLC patients. The results of analysis suggested SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in adenocarcinoma (ADC) whereas SIRT2 and SIRT5 were highly expressed in squamous cell carcinoma (SCC; ). SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in female patients whereas SIRT5 was highly expressed in male patients. High SIRT7 expression was associated with younger age at onset (<70 years vs ≥70 years). High SIRT1 expression was associated with distant metastasis (M1 vs M0) and low T stage (T1 vs T2–4). High SIRT4 was associated with high T stage (T2–4 vs T1) and TNM stage (II–IV vs I). High SIRT1 and SIRT4 was associated with non-smoking whereas high SIRT5 was associated with smoking.
Table 1 Correlations of sirtuins and clinicopathological parameters in patients with NSCLC
Correlations of sirtuins with prognosis in NSCLC patients
The correlations of sirtuins mRNA expressions with prognosis in NSCLC patients were investigated by Kaplan–Meier plot curves and Cox proportional regression analyses, wherein high SIRT2, SIRT4, and SIRT6 expressions were found to be associated with longer OS whereas no significant association of OS with other sirtuins was observed ( and ). Multivariate analyses indicated that SIRT2 and SIRT6 was still significantly associated with OS after adjusting for potential confounding factors including age, M, N, and T stages. For RFS, univariate analyses suggested high SIRT1 expression was significantly associated with shorter RFS time whereas high expressions of SIRT2, SIRT3, SIRT5, SIRT6, and SIRT7 were significantly associated with longer RFS time ( and ). After adjusting for the confounding factors, including histology, N, and T stages, significant associations were still found in SIRT1, SIRT2, SIRT5, SIRT6, and SIRT7 but not in SIRT3.
Table 2 Cox proportional regression analysis to investigate the correlations of sirtuins and overall survival in patients with NSCLC
Table 3 Cox proportional regression analysis to investigate the correlations of sirtuins and recurrence-free survival in patients with NSCLC
Figure 1 Overall survival in TCGA NSCLC patients with high and low sirtuin expression (SIRT1–7) were compared with Kaplan–Meier plot curves.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7.
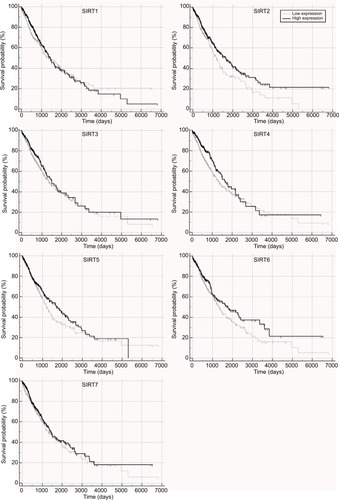
Figure 2 Recurrence-free survival in TCGA NSCLC patients stratified by high and low sirtuin expression (SIRT1–7) or a combination of SIRT1 and SIRT2 were compared with Kaplan–Meier plot curves. (A) SIRT1, (B) SIRT2, (C) SIRT3, (D) SIRT4, (E) SIRT5, (F) SIRT6, (G) SIRT7, and (H) Combination of SIRT1 and SIRT2.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; +, high expression; –, low expression.
Prediction of RFS with the combination of SIRT1 and SIRT2
Considering that SIRT1 and SIRT2 were the most significant biomarkers that were associated with RFS among the seven sirtuins and SIRT1 was a poor prognostic factor while SIRT2 was a favorable factor, we stratified the patients by combining SIRT1 and SIRT2 and then compared their RFS in order to provide a better prediction model for RFS in NSCLC. Kaplan–Meier plot curves and Cox proportional analyses suggested that, compared to patients with the SIRT1−/SIRT2+ genotype, patients with SIRT1−/SIRT2− or SIRT1+/SIRT2+ genotypes had poor RFS and patients with the SIRT1+/SIRT2− genotype had poorer prognosis ( and ).
Table 4 Cox proportional regression analysis to investigate the correlations of the combination of SIRT1 and SIRT2 with recurrence-free survival in patients with NSCLC
Integrated analysis of TCGA dataset and other publicly available datasets
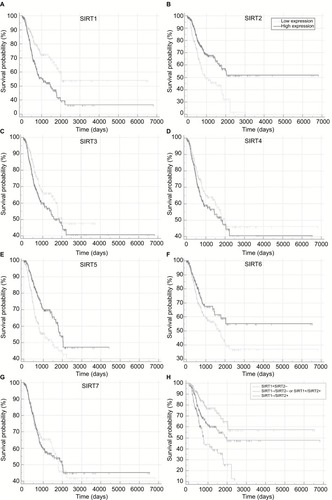
To further validate our results, we conducted an integrated analysis by pooling the TCGA NSCLC dataset and other public datasets. The Kaplan–Meier plotter (KM plotter) is an online tool to analyze correlations of individual genes with prognosis of patients in previous microarray datasets that comprised a total of 2,437 lung cancer cases. HRs with 95% CIs of the associations between sirtuins and RFS from each separate dataset in the database were obtained. After pooling these results by a meta-analysis method, high SIRT2 and SIRT7 expressions were found to be associated with favorable RFS in lung cancer ().
Figure 3 Forest plot for the correlations of sirtuins (SIRT1–7) and recurrence-free survival in patients with NSCLC by pooling the results of the datasets from TCGA and Kaplan–Meier plotter databases.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7.
Prognostic value of sirtuins in NSCLC reported in the published literature
Some articles have reported the associations of sirtuin expressions with prognosis in NSCLC patients. Here, we conducted a systematic literature search in three English databases (PubMed, EMBASE, and ISI Web of science) up to January 2018. In total, 14 articles published from 2013 to 2017 were identified after screening (), of which seven, two, two, one, and three articles reported the associations of SIRT1,Citation28–Citation34 SIRT2,Citation30,Citation36 SIRT3,Citation37,Citation38 SIRT5,Citation39 and SIRT6,Citation40–Citation42 respectively, with OS in NSCLC. Moreover, no article has reported SIRT4 and SIRT7 associations. High SIRT1 expression was found to be a poor prognostic factor in four reports, and no association was found in the remaining three articles. High SIRT2 was found to be associated with longer OS time in one article and high expressions of SIRT3, SIRT5, and SIRT6 were associated with poor OS. However, these reports were rare and their sample size was limited (SIRT3: N=2, n=133; SIRT5, N=1, n=39; SIRT6, N=3, n=394). Notably, the results for SIRT6 in the previous reports and our study were inconsistent, which needs to be clarified with a large sample size in the future.
Table 5 Associations of high sirtuins expressions with overall survival in NSCLC in previous reports
Analyses of the genes involved in NSCLC during sirtuin alteration
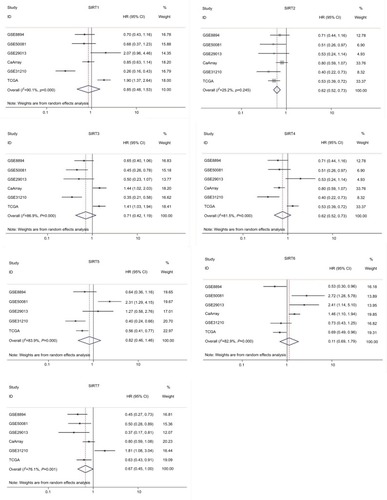
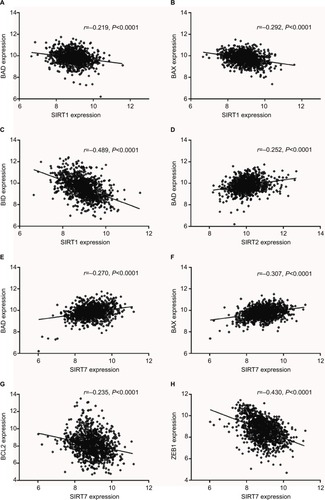
According to our screening of the literature, we found 18 articles reported the underlying mechanisms of sirtuins in lung cancer (). There were five, five, four, one, and four articles exploring the molecular mechanisms of SIRT1, SIRT2, SIRT3, SIRT5, and SIRT6 in lung cancer, respectively, whereas SIRT4 and SIRT7 were not investigated. To further understand the potential mechanisms of sirtuins in NSCLC carcinogenesis, genes that were positively or negatively associated with sirtuin expressions were analyzed and annotated. Pearson’s correlation analyses were conducted to obtain genes that were co-expressed with sirtuins in the 1017 TCGA NSCLC patients. The top 500 significant genes were selected and subjected to GO analysis with the GeneCoDis3 online tool (http://genecodis.cnb.csic.es). SIRT1, SIRT2, and SIRT7 were used as examples. Our results suggest that the top enriched biological processes were: regulation of transcription, signal transduction, apoptotic process, multicellular organismal development, positive regulation of cell proliferation, small GTPase-mediated signal transduction, and cell adhesion for SIRT1 (); regulation of transcription, apoptotic process, protein phosphorylation, RNA splicing, innate immune response, nerve growth factor receptor signaling, and vesicle-mediated transport for SIRT2 (); and signal transduction, DNA repair, protein phosphorylation, blood coagulation, nervous system development, small GTPase-mediated signal transduction, and cell proliferation for SIRT7 (). Cell apoptotic or proliferation processes are overlapping processes and have been reported to be involved in the mechanisms of SIRT1 and SIRT2 in NSCLC. Thereafter, the associations of SIRT1, SIRT2, and SIRT7 with the key genes in the apoptotic process including BAD, BAX, BCL2, and BID were investigated. The results suggested that SIRT1 was negatively associated with proapoptotic factors BAD, BAX, and BID whereas SIRT2 was positively associated with the proapoptotic factor, BAD (). Furthermore, SIRT7 was positively associated with proapoptotic factors BAD and BAX, and negatively associated with the antiapoptotic factor BCL2 (). In addition, SIRT7 was negatively asso ciated with ZEB1, a pro-proliferative factor (). To some extent, this explained the different functions of SIRT1, SIRT2, and SIRT7 in NSCLC carcinogenesis.
Figure 4 Biological processes regulated by SIRT1, SIRT2, and SIRT7 in TCGA NSCLC patients: (A) SIRT1, (B) SIRT2, and (C). SIRT7.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT7, sirtuin 7.
Figure 5 Expression correlations of SIRT1, SIRT2, and SIRT7 with BAD, BAX, BID, BCL2, and ZEB1 in TCGA NSCLC patients. (A–C). Correlations of SIRT1 with BAD (A), BAX (B), and BID (C). (D) Correlation of SIRT2 with BAD. (E–H) Correlations of SIRT7 with BAD (E), BAX (F), BCL2 (G), and ZEB1 (H).
Abbreviations: SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT7, sirtuin 7; BAX, Bcl-2 associated X; BAD, Bcl-2 associated agonist of cell death; BID, BH3 interacting domain death agonist; ZEB1, zinc finger E-box binding homeobox 1; TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer.
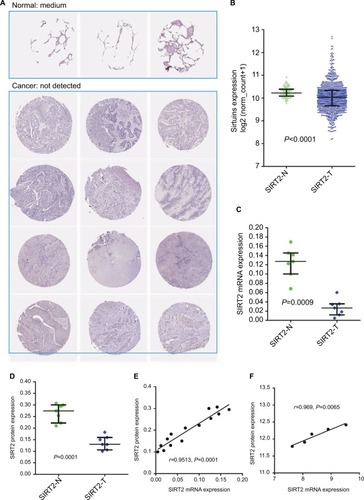
SIRT2 protein expression was in concordance with its mRNA expression in NSCLC
Genes generally function at the protein level; therefore, we investigated the concordance between sirtuin mRNA and protein expressions in NSCLC, with SIRT2 as an example. SIRT2 protein and mRNA expressions were initially compared between NSCLC tissues and corresponding controls. IHC staining data from the HPA suggested that SIRT2 protein expression was downregulated in NSCLC tissues compared with normal tissues (). The TCGA NSCLC data revealed that the SIRT2 mRNA expression was downregulated in NSCLC tissues (). In addition, we re-analyzed the results from Li et al’s study,Citation44 which detected SIRT2 mRNA expression by qRT-PCR, and protein expression by Western blotting in the same NSCLC patients; moreover, we found SIRT2 mRNA and protein expressions were downregulated in lung cancers compared with paired normal tissues (). Then, the correlation of SIRT2 mRNA and protein expressions was examined and a high positive correlation was found (r=0.9513, P<0.0001; ). Furthermore, we identified one study reporting both NSCLC proteomics and transcription profiles from the PRIDE databaseCitation46 and also found a high concordance between SIRT2 mRNA and protein expressions (r=0.9689, P=0.0065; ). The above results revealed that SIRT2 protein and mRNA expressions shared high concordance in NSCLC.
Figure 6 Concordance of SIRT2 mRNA and protein expressions in NSCLC. (A). SIRT2 immunohistochemical staining results in normal lung tissues (n=3) and lung cancer tissues (n=12). The images were obtained from the Human Protein Atlas.Citation43 (B). SIRT2 mRNA expression in normal lung tissues (n=110) and lung cancer tissues (n=1017) in the TCGA NSCLC cohort. (C–E). SIRT2 mRNA expression detected by qRT-PCR (C) and protein expression detected by Western blotting (D) in seven lung cancers and paired adjacent normal tissues (n=7); consistency of SIRT2 mRNA and protein expression were explored by Pearson’s correlation analysis (E). These were re-analyses results of Li et al’s report.Citation44 (F). Associations of SIRT2 mRNA with protein expressions in lung cancers. SIRT2 protein expression and mRNA expression data were extracted from proteomics and gene microarray profiles, respectively, which was reported by Stewart et al.Citation46
Abbreviations: SIRT2, sirtuin 2; N, normal; T, tumor; qRT-PCR, quantitative real-time polymerase chain reaction.
Discussion
Currently, a large number of cell signaling pathways have been discovered as being involved in tumor carcinogenesis and may serve as potential therapeutic targets for cancer treatment. Sirtuins are a family of mammalian proteins homologous to yeast Sir2.Citation6,Citation7 Seven different sirtuins (SIRT1–7) have been found and widely expressed in mammals. Previous reports have revealed the involvement of sirtuins in numerous critical biological processes including cell metabolism, genomic stability, cell cycle, cell division, differentiation, and transcriptional regulation and their functions in the pathogenesis of a variety of diseases such as metabolic diseases,Citation22 neurodegenerative diseases,Citation23 aging,Citation24 cardiovascular diseases,Citation25 and cancers.Citation26,Citation27 The clinical significance and potential molecular mechanisms of some sirtuins have been explored in lung cancer. However, the results are not consistent, the sample sizes in the previous studies are limited, and not all of the seven sirtuins have been explored and compared.Citation28–Citation42
In the present study, we utilized the mRNA profile dataset of 1017 NSCLC patients from the TCGA database to investigate and compare associations of the mRNA levels of the seven sirtuins (SIRT1–7) with clinicopathological parameters and prognosis in NSCLC. SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in ADC and female patients whereas SIRT5 was highly expressed in SCC and male patients. Associations of high SIRT7 with younger age at onset, high SIRT1 with distant metastasis and low T stage, and high SIRT4 with high T stage and TNM stage were also found. Furthermore, the prognostic value of sirtuins in NSCLC was investigated, and the results indicated that high mRNA levels of SIRT2 and SIRT6 were associated with longer OS duration. Moreover, high SIRT1 expression was significantly associated with shorter RFS time whereas high SIRT2, SIRT5, SIRT6, and SIRT7 expressions were associated with longer RFS time.
To compare and validate our results with that from previous reports, we undertook a systematic literature search to identify the full-text of original articles on the associations of sirtuins with OS in NSCLC (). SIRT1 was the most studied sirtuin for evaluating the associations of sirtuins with OS in lung cancer. A significant association of SIRT1 with OS was found in four of the seven reports.Citation28–Citation34 A recent meta-analysis has pooled the results from the above seven reports and found that high SIRT1 expression was a poor factor for OS in NSCLC,Citation35 whereas no significant association of SIRT1 with OS was found in our present study. This inconsistency might be attributed to the detection methods of SIRT1 and selection of cutoff value. Moreover, a discrepancy was found for the association of SIRT6 with OS. Three reports, with a total of 394 cases, suggested high SIRT6 expression was associated with poor OS, whereas our results suggested that high SIRT6 was associated with favorable OS.Citation40–Citation42 Until now, associations of SIRT4 or SIRT7 with prognosis of NSCLC patients have not been reported. More studies with large sample size should be conducted to clarify our results.
To date, some molecular mechanisms of sirtuins have been revealed in NSCLC (). SIRT1 can promote lung tumor growth through downregulating DLL4/Notch signaling and deacetylating the Notch1 intracellular domain;Citation47 its inactivation exerts antitumor activities through the tumor suppressor p27 in NSCLC;Citation48 and it is also involved in lung cancer development via the TNF-α/β-catenin axis under Benzo[a]pyrene (B[a]P) exposure – a carcinogen in cigarette smoke.Citation49,Citation50 SIRT2 can inhibit cell proliferation or induce cell apoptosis by increasing reactive oxygen species (ROS) production and p27 levels,Citation44 downregulating JMJD2A,Citation51 or deacetylating SKP2 followed by decreasing ubiquitination of p27.Citation36 SIRT3 can promote tumor proliferation and inhibit apoptosis via interaction with NMNAT2, and promote NSCLC malignancy via activation Akt at the post-transcriptional level.Citation38,Citation52 SIRT5 can promote cell proliferation and chemotherapy resistance by inducing Nrf2 expression.Citation39 An article reported that SIRT6 can increase tumor migration and invasion via enhancing ERK1/2 phosphorylation and MMP9 expression,Citation41 whereas another report suggested that SIRT6 can suppress cell proliferation through inhibition of Twist1 expression.Citation53 There is no report on the mechanisms of SIRT4 and SIRT7 in NSCLC. Modulation of cell proliferation and/ or apoptosis might be the common mechanisms of sirtuin action in NSCLC carcinogenesis. To further understand the potential mechanisms of sirtuins in NSCLC carcinogenesis, we investigated the cellular biological processes regulated by sirtuin alterations through annotating the sirtuin co-expressed genes by GO analysis. We took SIRT1, SIRT2, and SIRT7 as examples, because SIRT1 and SIRT2 were the mostly studied sirtuins and SIRT7 has not been investigated in NSCLC, and found they might affect tumor activities via modulating cell proliferation and apoptosis. The potential mechanisms explained the different prognostic value of SIRT1, SIRT2, and SIRT7. More in vitro and animal experiments should be undertaken to validate our findings.
Conclusion
Sirtuin expressions were significantly associated with clinicopathological parameters, especially tumor histology and gender, and clinical outcomes in NSCLC patients. For OS, high expressions of SIRT2 and SIRT6 were favorable prognostic factors. For RFS, high SIRT1 expression was a poor prognostic factor, whereas high SIRT2 and SIRT5–7 expressions were favorable prognostic factors. Combining SIRT1 and SIRT2 could provide a better prognostic model for RFS.
Acknowledgments
This study was supported by funding from the National Science Foundation for Young Scientists of China (grant no. 81601849) and Wenzhou Science and Technology Bureau (grant no. Y20160075).
Supplementary material
Table S1 Underlying mechanism of sirtuins in NSCLC in the previous reports
Disclosure
The authors report no conflicts of interest in this work.
References
- TorreLASiegelRLJemalALung cancer statisticsAdv Exp Med Biol201689311926667336
- TravisWDPathology of lung cancerClin Chest Med201132466969222054879
- ChenWZhengRZhangSZhaoPZengHZouXReport of cancer incidence and mortality in China, 2010Ann Transl Med2014276125333036
- PaoWGirardNNew driver mutations in non-small-cell lung cancerLancet Oncol201112217518021277552
- ParkSJMoreSMurtuzaAWoodwardBDHusainHNew targets in non-small cell lung cancerHematol Oncol Clin North Am201731111312927912827
- DonmezGOuteiroTFSIRT1 and SIRT2: emerging targets in neurodegenerationEMBO Mol Med20135334435223417962
- InoueTHiratsukaMOsakiMOshimuraMThe molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulationCell Cycle2007691011101817457050
- ImaiSArmstrongCMKaeberleinMGuarenteLTranscriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylaseNature2000403677179580010693811
- LandryJSuttonATafrovSTThe silencing protein SIR2 and its homologs are NAD-dependent protein deacetylasesProc Natl Acad Sci U S A200097115807581110811920
- NorthBJMarshallBLBorraMTDenuJMVerdinEThe human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylaseMol Cell200311243744412620231
- TannerKGLandryJSternglanzRDenuJMSilent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-riboseProc Natl Acad Sci U S A20009726141781418211106374
- HaigisMCMostoslavskyRHaigisKMSIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cellsCell2006126594195416959573
- LisztGFordEKurtevMGuarenteLMouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferaseJ Biol Chem200528022213132132015795229
- BarberMFMichishita-KioiEXiYSIRT7 links H3K18 deacetylation to maintenance of oncogenic transformationNature2012487740511411822722849
- MichishitaEMcCordRABoxerLDCell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6Cell Cycle20098162664266619625767
- KumarSLombardDBFunctions of the sirtuin deacylase SIRT5 in normal physiology and pathobiologyCrit Rev Biochem Mol Biol201853331133429637793
- DuJZhouYSuXSirt5 is a NAD-dependent protein lysine demalonylase and desuccinylaseScience2011334605780680922076378
- ParkJChenYTishkoffDXSIRT5-mediated lysine desuccinylation impacts diverse metabolic pathwaysMol Cell201350691993023806337
- NishidaYRardinMJCarricoCSIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major targetMol Cell201559232133226073543
- TanMPengCAndersonKALysine glutarylation is a protein posttranslational modification regulated by SIRT5Cell Metab201419460561724703693
- LiLShiLYangSSIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stabilityNat Commun201671223527436229
- HoutkooperRHPirinenEAuwerxJSirtuins as regulators of metabolism and healthspanNat Rev Mol Cell Biol201213422523822395773
- BondaDJLeeHGCaminsAThe sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerationsLancet Neurol201110327527921349442
- HallJADominyJELeeYPuigserverPThe sirtuin family’s role in aging and age-associated pathologiesJ Clin Invest2013123397397923454760
- BorradaileNMPickeringJGNAD(+), sirtuins, and cardiovascular diseaseCurr Pharm Des200915111011719149606
- Bosch-PreseguéLVaqueroAThe dual role of sirtuins in cancerGenes Cancer20112664866221941620
- YuanHSuLChenWYThe emerging and diverse roles of sirtuins in cancer: a clinical perspectiveOnco Targets Ther201361399141624133372
- NohSJBaekHAParkHSExpression of SIRT1 and cortactin is associated with progression of non-small cell lung cancerPathol Res Pract2013209636537023702379
- ZhangTRongNChenJSIRT1 expression is associated with the chemotherapy response and prognosis of patients with advanced NSCLCPLoS One2013811e7916224223900
- GrbesaIPajaresMJMartínez-TerrobaEExpression of sirtuin 1 and 2 is associated with poor prognosis in non-small cell lung cancer patientsPLoS One2015104e012467025915617
- LiCWangLZhengLSIRT1 expression is associated with poor prognosis of lung adenocarcinomaOnco Targets Ther2015897798425995644
- GharabaghiMADiagnostic investigation of BIRC6 and SIRT1 protein expression level as potential prognostic biomarkers in patients with non-small cell lung cancerClin Respir J201812263363827768839
- LinSYPengFAssociation of SIRT1 and HMGA1 expression in non-small cell lung cancerOncol Lett201611178278826834854
- WangJWangCPrognostic and predictive role of sirtuin1 expression in lung adenocarcinomaClin Lab201662101989199428164517
- ChenYWangTWangWPrognostic and clinicopathological significance of SIRT1 expression in NSCLC: a meta-analysisOncotarget2017837625376254428977967
- LiZHuangJYuanHChenZLuoQLuSSIRT2 inhibits non-small cell lung cancer cell growth through impairing Skp2-mediated p27 degradationOncotarget2016714189271893926942878
- YangGCFuBCZhangDYThe expression and related clinical significance of SIRT3 in non-small-cell lung cancerDis Markers201720178241953
- XiongYWangMZhaoJSIRT3 is correlated with the malignancy of non-small cell lung cancerInt J Oncol201750390391028197634
- LuWZuoYFengYZhangMSIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancerTumour Biol20143511106991070525070488
- AzumaYYokoboriTMogiASIRT6 expression is associated with poor prognosis and chemosensitivity in patients with non-small cell lung cancerJ Surg Oncol2015112223123726180037
- BaiLLinGSunLUpregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathwayOncotarget2016726403774038627777384
- ChenTSunZLiuFWangQRASSF1A and SIRT6 in non-small cell lung cancer: relationship with clinical outcomeOncol Lett20171455759576429204192
- UhlenMOksvoldPFagerbergLTowards a knowledge-based Human Protein AtlasNat Biotechnol201028121248125021139605
- LiZXieQRChenZLuSXiaWRegulation of SIRT2 levels for human non-small cell lung cancer therapyLung Cancer201382191523915912
- LiLWeiYToCIntegrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impactNat Commun20145546925429762
- StewartPAParapaticsKWelshEAA pilot proteogenomic study with data integration identifies MCT1 and GLUT1 as prognostic markers in lung adenocarcinomaPLoS One20151011e014216226539827
- XieMLiuMHeCSSIRT1 regulates endothelial Notch signaling in lung cancerPLoS One201279e4533123028940
- ZhuLChiaoCYEnzerKGStankiewiczAJFallerDVDaiYSIRT1 inactivation evokes antitumor activities in NSCLC through the tumor suppressor p27Mol Cancer Res2015131414925143434
- LuJZhangMHuangZSIRT1 in B[a]P-induced lung tumorigenesisOncotarget2015629271132712926318035
- HuangHPanXJinHPHLPP2 downregulation contributes to lung carcinogenesis following B[a]P/B[a]PDE exposureClin Cancer Res201521163783379325977341
- XuWJiangKShenMQianYPengYSIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2ABiol Chem2015396892993625719312
- LiHFengZWuWLiJZhangJXiaTSIRT3 regulates cell proliferation and apoptosis related to energy metabolism in non-small cell lung cancer cells through deacetylation of NMNAT2Int J Oncol20134351420143024042441
- HanZLiuLLiuYLiSSirtuin SIRT6 suppresses cell proliferation through inhibition of Twist1 expression in non-small cell lung cancerInt J Clin Exp Pathol20147847744781 eCollection 201425197348
- HanLLiangXHChenLXBaoSMYanZQSIRT1 is highly expressed in brain metastasis tissues of non-small cell lung cancer (NSCLC) and in positive regulation of NSCLC cell migrationInt J Clin Exp Pathol201361123572365 eCollection 201324228097
- ChengDZhaoLXuYK-Ras promotes the non-small lung cancer cells survival by cooperating with sirtuin 1 and p27 under ROS stimulationTumour Biol20153697221723225894374
- LuoJBaoYCJiXXChenBDengQFZhouSWSPOP promotes SIRT2 degradation and suppresses non-small cell lung cancer cell growthBiochem Biophys Res Commun2017483288088428073696
- XiaoKJiangJWangWSirt3 is a tumor suppressor in lung adenocarcinoma cellsOncol Rep20133031323132823842789
- XiongYWangLWangSSIRT3 deacetylates and promotes degradation of P53 in PTEN-defective non-small cell lung cancerJ Cancer Res Clin Oncol2018144218919829103158
- KimEJJuhnnYSCyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathwayJ Biol Chem2015290159604961325713071
