Abstract
Background
The aim of the current study was to investigate the potential prognostic value of minichromosome maintenance (MCM) genes in patients with early-stage pancreatic ductal adenocarcinoma (PDAC) after pancreaticoduodenectomy by using the RNA-sequencing dataset from The Cancer Genome Atlas (TCGA).
Methods
An RNA-sequencing dataset of 112 early-stage PDAC patients who received a pancreaticoduodenectomy was obtained from TCGA. Survival analysis was used to identify potential prognostic values of MCM genes in PDAC overall survival (OS).
Results
Through mining public databases, we observed that MCM genes (MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7) were upregulated in pancreatic cancer tumor tissue and have a strong positive coexpression with each other. Multivariate survival analysis indicated that a high expression of MCM4 significantly increased the risk of death in patients with PDAC, and time-dependent receiver operating characteristic analysis showed an area under the curve of 0.655, 0.587, and 0.509 for a 1-, 2-, and 3-year PDAC OS prediction, respectively. Comprehensive survival analysis of MCM4 using stratified and joint effects survival analysis suggests that MCM4 may be an independent prognostic indicator for PDAC OS. Gene set enrichment analysis indicated that MCM4 may participate in multiple biologic processes and pathways, including DNA replication, cell cycle, tumor protein p53, and Notch signaling pathways, thereby affecting prognosis of PDAC patients.
Conclusions
Our study indicates that MCM2–7 were upregulated in pancreatic cancer tumor tissues, and mRNA expression of MCM4 may serve as an independent prognostic indicator for PDAC OS prediction after pancreaticoduodenectomy.
Introduction
Pancreatic cancer (PC) is not only a highly lethal malignancy but also has a poor prognosis, low early diagnosis rate, and lacks effective treatment strategies.Citation1–Citation3 In 2012, it was estimated that there would be approximately 330,700 PC deaths worldwide.Citation4 In 2015, the National Central Cancer Registry of China (NCCRC) estimated that there would be approximately 901,000 new cases of PC and 794,000 deaths in China, using data from population-based cancer registries (2009–2011).Citation5 The updated nationwide cancer statistics from the NCCRC, which were estimated on population-based cancer registry data collected from all available cancer registries in 2014, suggest an upward trend in new cases of PC and deaths in China compared to their previous report (2009–2011).Citation5,Citation6
Due to the clinicopathologic features of PC, treatment and management strategies of PC were focused on developing early diagnostic and prognostic monitoring biomarkers. Previous studies have demonstrated that genetic alterations may have a contribution in PC tumorigenesis and progression.Citation7,Citation8 More than 80% of the histologic type of PCs are pancreatic ductal adenocarcinoma (PDAC).Citation4,Citation9,Citation10 Numerous studies have reported that minichromosome maintenance (MCM) genes (including MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7) could serve as a diagnostic and prognostic indicator in multiple cancers.Citation11–Citation14 However, the potential application value and mechanism of MCM genes in the prognostic prediction of PDAC after pancreaticoduodenectomy remain unclear. Therefore, the purpose of the current study was to investigate the potential prognostic application of MCM2–7 for patients with early-stage PDAC after pancreaticoduodenectomy by using an RNA-sequencing (RNA-Seq) dataset from The Cancer Genome Atlas (TCGA).
Materials and methods
Public database mining of MCM genes
With the development of high-throughput technologies, a large number of genome-wide expression profiling data have been published and freely shared with researchers, especially the whole genome dataset of cancer diseases. In the present study, we used the Genotype-Tissue Expression (GTEx; https://www.gtexportal.org/home/, accessed March 20, 2018) website to investigate the mRNA expression distribution of MCM genes in normal organ tissues.Citation15–Citation17 The Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html; accessed March 20, 2018) website was used to investigate the mRNA expression distribution of MCM genes in pancreatic adenocarcinoma (PAAD), which is an online analysis tool based on the RNA-Seq expression data of 9,736 tumors and 8,587 normal samples from TCGA and the GTEx projects.Citation18 We also used the Metabolic gEne RApid Visualizer (MERAV; http://merav.wi.mit.edu/; accessed March 20, 2018), comprising human gene expression data from normal tissues, primary tumors, and cancer cell lines, to further validate the expression distribution of MCM genes between PC and adjacent normal tissues.Citation19
Comprehensive survival analysis of MCM genes
The RNA-Seq dataset of PAAD was downloaded from TCGA (https://portal.gdc.cancer.gov/; accessed April 20, 2017)Citation20 and the raw data were normalized by DESeq.Citation21 The corresponding clinical information was downloaded from the University of California, Santa Cruz Xena (UCSC Xena: http://xena.ucsc.edu/, accessed April 20, 2017). To ensure the reliability of prognostic analysis, we have established a strict patient inclusion criteria and exclusion criteria, which have been published in our previous paper and are described as follows: 1) complete survival data available; 2) the histology type was PDAC; 3) pathologic stage I or II; 4) patients underwent pancreaticoduodenectomy.Citation22 PDAC patients with pathologic stage III or IV disease who underwent other types of surgery were excluded.Citation22 To comprehensively investigate the prognostic values of MCM genes in PDAC, multivariate Cox proportional risk regression model analysis was used to screen the genes by adjusting for clinical parameters that were significantly associated with PDAC overall survival (OS) in univariate survival analysis. Subsequently, comprehensive survival analysis of prognostic-MCM genes was performed, including evaluation by the survivalROC package in the R platform, stratified survival analysis, nomogram construction, and joint effect survival analysis.
Gene set enrichment analysis
To investigate the biologic processes involved in the clinical outcome of PDAC with different expression levels of prognostic-MCM genes, we carried out a gene set enrichment analysis (GSEA, http://software.broadinstitute.org/gsea/index.jsp, accessed December 15, 2017).Citation23,Citation24 GSEA was used to deep mine the potential mechanisms of prognostic-MCM genes using the Molecular Signatures Database (MSigDB) c2 (c2.cp.kegg.v6.1.symbols.gmt) and c5 (c5.all.v6.1.symbols.gmt).Citation25 The significantly enriched gene sets in the GSEA were identified with the following criteria: a nominal P-value <0.05 and false discovery rate (FDR) <0.25.
Statistical analysis
FDRs in the GSEA were adjusted for multiple testing with the Benjamini–Hochberg procedure to control FDR.Citation26–Citation28 Univariate survival analysis of clinical features and MCM genes were compared using the log-rank test; clinicopathologic parameters significantly associated with OS (P<0.05) were entered into the multivariate Cox proportional hazards regression model for adjustment, whereas, hazard ratios (HRs) and 95% CIs were used to assess the relative risk in different PDAC patients that were stratified by MCM gene expression. Coexpression relationships between MCM genes were assessed by the Pearson correlation coefficient. All statistical analyses were conducted with SPSS version 20.0 (IBM Corporation, Armonk, NY, USA) and R3.3.0. Statistical significance was set as a P-value <0.05.
Results
Public database mining of MCM genes
To make a complete investigation of public databases to analyze the distribution and function of MCM genes in humans, the distribution of MCM genes in human normal organ tissues was investigated by GTEx and is shown in . The gene distribution suggests that the expression of MCM2–7 genes was low in human pancreas tissues compared to other organ tissues. An investigation using GEPIA indicated that MCM2–7 genes were upregulated in human tumor tissues (Figure S1A–F and Figure S2 A–F), which is based on TCGA and GTEx databases, as well as in PAAD (). The distribution of upregulated MCM2–7 genes in PC was also verified by the MERAV online analysis tool (), which is based on the database of Gene Expression Omnibus.
Figure 1 Expression distribution of MCM2–7 in human normal organ tissues.
Notes: Expression distribution box plot of MCM2 (A); MCM3 (B); MCM4 (C); MCM5 (D); MCM6 (E); MCM7 (F).
Abbreviation: MCM, minichromosome maintenance.
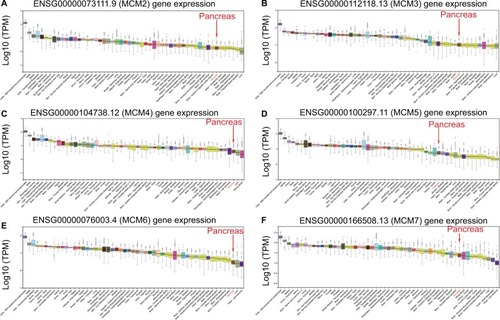
Figure 2 Expression distribution of MCM2–7 between PAAD tumor and normal tissues.
Notes: Expression distribution box plot of MCM2 (A); MCM3 (B); MCM4 (C); MCM5 (D); MCM6 (E); MCM7 (F). Expressions of MCM genes were upregulated in PDAC tumor tissues. *P<0.05.
Abbreviations: MCM, minichromosome maintenance; PAAD, pancreatic adenocarcinoma.
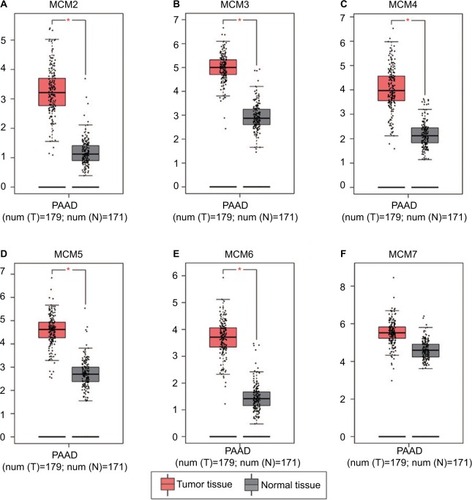
Figure 3 Expression distribution of MCM2–7 between PC tumor and normal tissues that was carried out by MERAV. Expressions of MCM genes were upregulated in PC tumor tissues.
Notes: Expression distribution box plot of MCM2 (A); MCM3 (B); MCM4 (C); MCM5 (D); MCM6 (E); MCM7 (F).
Abbreviations: MCM, minichromosome maintenance; MERAV, Metabolic gEne RApid Visualizer; PC, pancreatic cancer.
Bioinformatics analysis
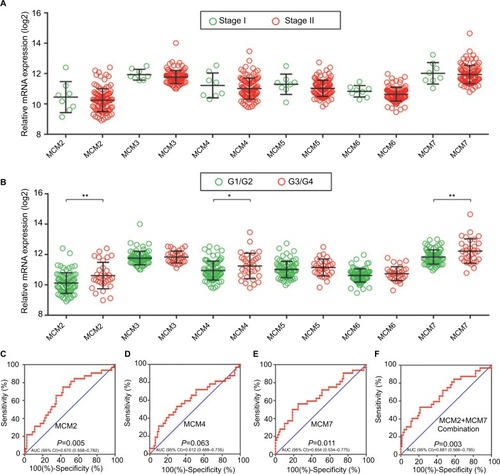
The RNA-Seq dataset of early-stage PDAC patients who underwent pancreaticoduodenectomy was obtained from TCGA PAAD project.Citation22 By implementing inclusion and exclusion criteria, a total of 112 early-stage PDAC patients met the above criteria and were included in further analysis. Coexpression of MCM2–7 genes in PDAC tumor tissue is shown in , which substantiates the strongly positive coexpression of these genes in PDAC tumor tissues (Pearson correlation coefficient ranged 0.220–0.745; all P<0.05). The strongly positive coexpression of these genes could also be observed in normal human pancreatic tissue in GEPIA, which is based on the GTEx database (, Pearson correlation coefficient ranged 0.280–0.780; all P<0.05). The bioinformatics prediction performed by Gene Multiple Association Network Integration Algorithm (GeneMANIA, http://www.genemania.org/, accessed March 20, 2018)Citation29 and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, https://string-db.org/, accessed March 20, 2018)Citation30 also suggest that MCM2–7 genes were strongly coexpressed with each other and with complex gene–gene and protein–protein interaction networks (Figure S3A, B). In addition, we investigated the expression distribution of MCM2–7 genes in different tumor stages and histologic grades and found that the expression of MCM2–7 genes was not significantly differentially distributed between stage I and stage II tumor tissues (), whereas expressions of MCM2, MCM4, and MCM7 were significantly differentially distributed between histologic grade G1/G2 and G3/G4 tumor tissues (). Furthermore, we also explored the ability of MCM2, MCM4, and MCM7 () to distinguish histologic grade G1/G2 and G3/G4 tumor tissues using the area under the curve (AUC) of a receiver operating characteristic (ROC) curve. ROC analysis indicated that MCM2 (P=0.005, AUC=0.670, 95% CI=0.558–0.782; ) and MCM7 (P=0.011, AUC=0.654, 95% CI=0.534–0.775; ) may have potential in distinguishing histologic grade G1/G2 and G3/G4 tumor tissues in PDAC, and a combination of MCM2 and MCM6 could improve diagnostic efficiency (P=0.003, AUC=0.681, 95% CI=0.566–0.795; ).
Figure 4 Coexpression matrix of MCM genes in PDAC tumor tissues and human normal pancreas tissues, and demonstrated that MCM2–7 were strongly positively correlation and coexpressed with each other in both PDAC tumor and normal pancreas tissues.
Notes: (A) Coexpression matrix of MCM genes in PDAC tumor tissues; (B) coexpression matrix of MCM genes in human normal pancreas tissues.
Abbreviations: MCM, minichromosome maintenance; PDAC, pancreatic ductal adenocarcinoma.
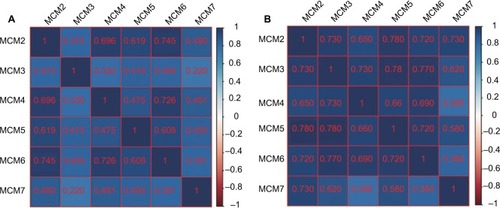
Figure 5 Gene expression distribution of MCM genes in different PDAC tumor stages and histologic grades, and ROC curves of MCM genes to distinguish different PDAC histologic grades.
Notes: (A) Gene expression distribution of MCM genes in different PDAC tumor stages; (B) gene expression distribution of MCM genes in different PDAC histologic grades; ROC curves of MCM2 (C), MCM4 (D), MCM7 (E) and MCM2 + MCM7 (F) combination to distinguish different PDAC histological grades. *P<0.05, **P<0.01.
Abbreviations: AUC, area under the curve; MCM, minichromosome maintenance; PDAC, pancreatic ductal adenocarcinoma; ROC, receiver operating characteristic.
Survival analysis of MCM2–7 in PDAC OS
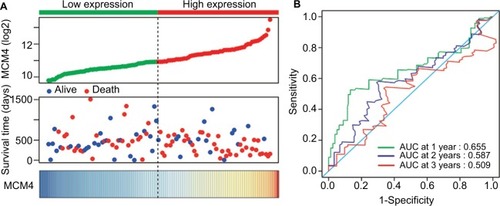
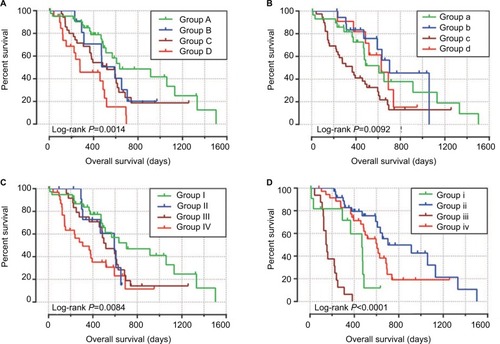
Baseline information of these 112 PDAC patients is shown in , and clinical parameters of histologic grade, radical resection, radiation therapy, and targeted molecular therapy were significantly associated with PDAC OS, which needed to be adjusted into a multivariate Cox proportional hazards regression model. Survival analysis of the MCM2–7 genes () demonstrated that MCM4 was significantly associated with PDAC OS, and high expression of MCM4 was significantly increased with the risk of death in PDAC (adjusted P=0.003, adjusted HR=2.409, 95% CI=1.351–4.294, , ) and promoted a poor OS (high MCM4 vs low MCM4; 458 days vs 634 days, , and ). We also performed a time-dependent ROC analysis, which was carried out using survivalROC in the R platform, to evaluate the predictive accuracy of MCM4 expression in PDAC OS, and demonstrated that MCM4 expression performed well in predicting PDAC OS. The AUC of the time-dependent ROC curve was 0.655, 0.587, and 0.509 for 1-, 2-, and 3-year survival (), respectively.
Table 1 Correlation between OS and clinicopathologic features of PDAC patientsTable Footnotea
Table 2 Prognostic values of MCM genes expression in PDAC OS of TCGA cohort
Figure 6 Kaplan–Meier survival curves for MCM genes in pancreatic ductal adenocarcinoma of The Cancer Genome Atlas cohort.
Notes: Overall survival stratified by MCM2 (A), MCM3 (B), MCM4 (C), MCM5 (D), MCM6 (E), and MCM7 (F).
Abbreviation: MCM, minichromosome maintenance.
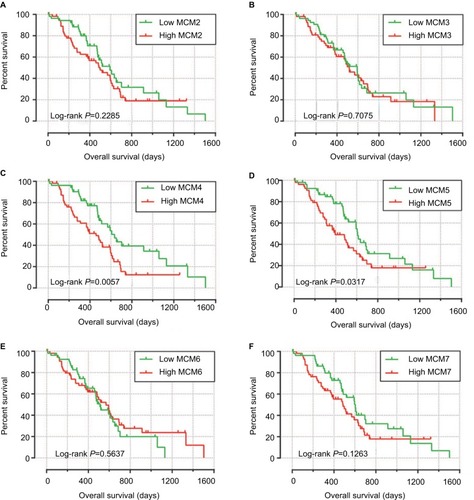
Figure 7 Prognostic value evaluation of MCM4 in patients with PDAC.
Notes: (A) From top to bottom are the expression values of MCM4, patients’ survival status distribution, and the expression heat map of MCM4 in the low- and high-expression groups. (B) Receiver operating characteristic curve for predicting overall survival in PDAC patients by the MCM4.
Abbreviations: MCM, minichromosome maintenance; PDAC, pancreatic ductal adenocarcinoma.
Comprehensive analysis of MCM4 in PDAC OS
To perform a comprehensive investigation of the role of MCM4 in the prognosis of PDAC, we used a stratified analysis and joint effect survival analysis to assess the prognostic value of MCM4 in PDAC, and developed a nomogram, which included clinical parameters and the MCM4 gene, to evaluate individualized prognostic risk scores. Stratified analysis suggests that high expression of MCM4 was associated with a significantly increased risk of death in PDAC patients except in patients with pathologic stage I, who received radiation therapy and targeted molecular therapy; young patients (≤60 years); female patients, without or with alcohol history; and patients without radical resection (). The nomogram indicated that expression of MCM4 also had a certain contribution to the prognosis of PDAC (). Joint effect survival analysis suggests that a combination of MCM4 and those clinical parameters, which were significantly correlated to PDAC OS, showed a better performance in PDAC OS, compared with single clinical parameters ( and ).
Table 3 Joint effects survival analysis of clinical factors and the MCM4 expression with OS in PDAC patients
Figure 8 The relationship between MCM4 and clinical information.
Notes: (A) Stratified analysis of association between MCM4 and overall survival in pancreatic ductal adenocarcinoma. (B) Nomogram for predicting the 1-, 2-, and 3-year event (death) with MCM4 and clinical information.
Abbreviation: MCM, minichromosome maintenance.
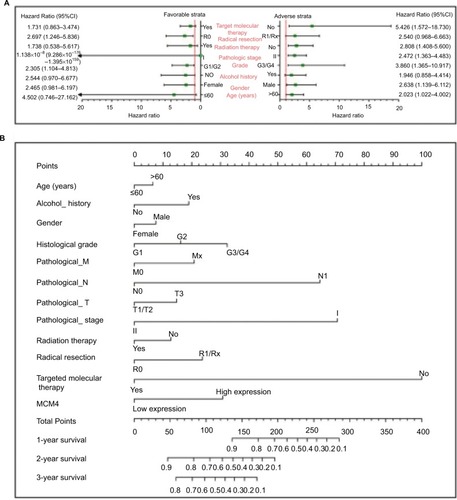
Figure 9 Joint effects analysis of overall survival stratified by MCM4 and pancreatic ductal adenocarcinoma clinical parameters.
Notes: Joint effects analysis stratified by MCM4 and following clinical parameters: Histologic grade (A), radiation therapy (B), radical resection (C), and targeted molecular therapy (D).
Abbreviation: MCM, minichromosome maintenance.
GSEA investigation for MCM4 in PDAC OS
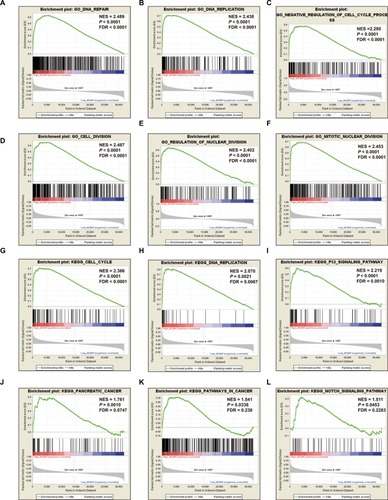
To further explore the potential mechanism of MCM4 in PDAC OS, we also developed a single-gene GSEA to investigate the potential biologic processes and pathways between different MCM4 expression levels. Enrichment of c5 suggests that high expression of MCM4 may be involved in DNA repair, DNA replication, cell cycle, and cell and nuclear division biologic processes (, Table S1), whereas enrichment of c2 indicates that high expression of MCM4 may participate in the cell cycle, DNA replication, tumor protein p53 (TP53), PC, and Notch signaling pathways, as well as pathways in cancer (; Table S2).
Figure 10 GSEA results of MCM4 in PDAC patients.
Notes: (A–F) GSEA results of c2 reference gene sets for high MCM4 expression groups; (G–L) GSEA results of c5 reference gene sets for high MCM4 expression groups.
Abbreviations: ES, enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; MCM, minichromosome maintenance; NES, normalized enrichment score; PDAC, pancreatic ductal adenocarcinoma.
Discussion
The replication of DNA is a fundamental step in the cell cycle, and the MCM protein family in this process has been investigated extensively in numerous studies over the past decade.Citation31–Citation35 The MCM2–7 complex provides essential replicative helicase function in late mitosis and early G1 in the cell cycle.Citation35–Citation37 The functions of MCM2–7 are involved in the basic biologic processes of cell cycle maintenance in cells; therefore, dysregulation of the MCM protein family may cause cancer or developmental defects.Citation38–Citation41 MCM genes may be potential diagnostic and prognostic biomarkers for cancers, and promising targets for anticancer drug development and targeted therapy.Citation42
Extensive studies have shown that the dysregulation of MCM2–7 can be observed in various cancers and may have a diagnostic value. Work by Saydam et al demonstrated that expression of MCM2–7 mRNAs was markedly increased in meningiomas tumor tissue.Citation43 A prospective cohort study from India suggests that MCM (MCM2 and MCM5) immunocytochemistry may have considerable advantages for first-line cervical screening in developing countries.Citation44 Immunohistochemical detection of MCM2 in the urine of bladder cancer (BC) patients and in the stools of colorectal cancer (CRC) patients could serve as a novel method for the diagnosis of these cancers.Citation45,Citation46 A study by Wang et al observed that mRNA expression of MCM2 in colon adenocarcinoma (COAD) was significantly increased in adenomas, as well as upregulation in adenomas with high-grade dysplasia, or in older patients, and demonstrates an application value in the early diagnosis of COAD.Citation47 Immunohistochemical detection of MCM5 in human secretions also has a diagnostic value for cancers of the corresponding organs, such as urine sediments for prostate cancer,Citation13 gastric aspirates for esophageal cancer,Citation14 and bile aspirates or biliary brush cytology for pancreaticobiliary malignancy.Citation48,Citation49 Moreover, the levels of MCM6 mRNA and protein in the plasma are also markedly increased, and significantly associated with tumor stage progression and lymph node metastasis in hepatocellular carcinoma (HCC).Citation50 Similar to previous studies, by using the GEPIA online analysis tool, we observed that MCM2–7 were upregulated in the majority of human tumor tissues, as well as in PC tumor tissue. However, due to the limitation that we cannot obtain normal pancreas tissue expression data from GEPIA, we could not assess the diagnostic values of MCM2–7 in PC.
In addition, extensive investigations indicate that MCM genes may serve as proliferation markers in various cancers. Immunohistochemical analysis of MCM3 might be useful as a proliferation marker in oral squamous cell carcinoma (OSCC),Citation51 papillary thyroid carcinoma,Citation52 and salivary gland tumors.Citation53 The potential application of MCM5 in identifying cell proliferation can be found in Merkel cell carcinomaCitation54 and malignant skin diseases.Citation55 Similar studies of MCM7 also have been reported in gastric cancer (GC),Citation56 esophageal lesions,Citation57 reactive mesothelial cells, and malignant cells.Citation58,Citation59 Because of the close relationship between MCM gene and cell proliferation, MCM genes are also found to be associated with tumor progression. Overexpression of MCM7 was significantly associated with prostate cancer progression, relapse, local invasion, and a worse tumor grade,Citation60 as well as in OSCC development and metastasis.Citation61 Work by Das et al observed that MCM4, MCM5, MCM6, and MCM10 were significantly overexpressed in cervical cancer tumor tissues, and upregulated in advanced tumor stages, which indicates that these genes were significantly associated with cervical cancer carcinogenesis and progression.Citation62 Moreover, there was a significantly positive association between the expression of MCM2 and histopathologic grade, and the expression was markedly upregulated in poorly differentiated HCC tissues, which may serve as a biomarker for HCC progression.Citation63
The prognostic values of MCM2–7 have also been identified in multiple types of cancer.Citation64–Citation66 Due to the upregulation of MCM2–7 in multiple types of cancer, we speculate that they might play an oncogene role in cancer prognosis, and high expression of MCM genes may promote a poor survival. The following series of evidence from a review of the literature will support our inference. Work by Kang et al also demonstrated that knockdown of MCM7 in GC cells may suppress its oncogenic function.Citation65 Immunohistochemical detection of MCM2 can be used as a biomarker for clinical outcome prediction, and high expression of MCM2 was associated with a significantly increased risk of death in patients with muscle-invasive urothelial cancer,Citation66 nonbenign epithelial ovarian tumors,Citation67 non-small-cell lung cancer (NSCLC),Citation12,Citation68 and GC,Citation69–Citation71 or recurrence in patients with BCCitation72 and GC.Citation71 Similar results could also be observed with MCM5, MCM6, and MCM7. High expression of MCM5 was associated with a significantly increased risk of death in patients with NSCLCCitation73 and cervical cancer,Citation74 and patients with high expression of MCM6 also have poor OS and increased risk of death in NSCLC,Citation75 low-grade chondrosarcoma,Citation76 mantle cell lymphoma,Citation77 and endometrioid endometrial adenocarcinoma.Citation78 The application value of MCM7 in the prognosis of cancer has been widely investigated. Previous studies have substantiated that MCM7 expression can serve as an independent prognostic factor for human CRC,Citation79 HCC,Citation80 lung cancer,Citation81,Citation82 Hodgkin’s lymphoma,Citation83 OSCC,Citation84 and esophageal squamous cell carcinoma,Citation85 and promotes a poor prognosis. In addition, MCM7 also plays a crucial role in monitoring recurrence or progression-free survival, and evidence from previous studies suggests that MCM7 can be used as a biomarker for recurrence of CRC,Citation86 meningiomas,Citation87 and GC,Citation65 as well as progression-free survival of non-muscle-invasive BC,Citation88 pituitary adenoma,Citation89 and ovarian cancer.Citation90 A previous study by Peng et al reported that MCM genes were significantly overexpressed in PC tumor tissue and correlated to PC progression and prognosis, which is based on PC patients from TCGA.Citation91 However, the study by Peng et al did not consider the influence of different operation methods and histologic subtypes for PC prognosis. The advantage of this current study was to take into full consideration the effect of operation method and histologic subtypes in PC prognosis, and only included patients with PDAC histologic subtype and underwent pancreaticoduodenectomy; therefore, the results obtained from our study may be more reliable. In addition, we also developed a comprehensive survival analysis for MCM4 and constructed a nomogram base on the clinical parameters and MCM4 mRNA expression levels, indicating that MCM4 may be an independent prognostic indicator for PDAC OS prediction after pancreaticoduodenectomy.
The exploration of a potential mechanism by GSEA revealed that MCM4 may take part in the biologic processes and pathways of DNA replication, cell cycle, TP53, and Notch signaling, which may affect PDAC prognosis. By reviewing the literature, we also found that a number of previous studies supported our results and substantiated that MCM4 played a crucial role in the cell cycle,Citation92,Citation93 DNA replication,Citation94,Citation95 DNA repair,Citation96 and TP53 signaling pathwayCitation97; these biologic processes and pathways have already demonstrated a role in cancer prognosis.
As all the data in the present study come from public databases, there are several limitations that need to be recognized. First, the clinical information from TCGA database was not comprehensive, and the results of survival analysis still need to be verified. Second, due to strict inclusion and exclusion criteria, the sample size in our study was relatively small; therefore, an additional large verification cohort to validate our results is necessary. Third, due to a relatively small sample size, and because most of the patients died within 3 years, we cannot assess the survival prediction accurately for more than 3 years. Fourth, because adjacent normal tissues of PDAC were rare, we cannot perform an ROC analysis to assess the diagnostic value of MCM2–7 in PDAC.
Despite these limitations, in the present study we have identified the prognostic application of MCM4 mRNA expression in patients with PDAC, and also investigated the potential mechanism of different MCM4 expression levels in PDAC prognosis through a GSEA approach. Once these results are confirmed, MCM4 may have potential in the clinical application of prognostic monitoring, cancer management, and targeted therapy of PDAC.
Conclusions
Through a comprehensive analysis of MCM genes, we observed that MCM2–7 were upregulated in PC tumor tissues, and mRNA expression of MCM4 may serve as an independent prognostic indicator for PDAC prognosis prediction. The potential mechanism of MCM4 in PDAC OS, which was investigated by GSEA, indicated that MCM4 may play a role in PDAC prognosis through participating in the biologic processes and pathways of DNA replication, cell cycle, TP53, and Notch signaling. However, these results still need further verification and investigation.
Acknowledgments
This work was supported in part by the National Nature Science Foundation of China (Nos.: 81560535, 81072321, 30760243, 30460143, and 30560133), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Nature Sciences Foundation (No.: GuiKeGong 1104003A-7), and Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018). The present study is also partly supported by Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (Z2016318) and The Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (2018KY0110). The present study is also partly supported by Research Institute of Innovative Think-tank in Guangxi Medical University (The gene–environment interaction in hepatocarcinogenesis in Guangxi HCCs and its translational applications in the HCC prevention). We would also acknowledge the support by the National Key Clinical Specialty Programs (General Surgery and Oncology) and the Key Laboratory of Early Prevention & Treatment for Regional High-Incidence-Tumor (Guangxi Medical University), Ministry of Education, China. The authors thank the contributors of TCGA (https://cancergenome.nih.gov/) and UCSC Xena (http://xena.ucsc.edu/) for sharing the PDAC data on open access. In addition, we would like to acknowledge the helpful comments on this article received from our reviewers.
Disclosure
The authors report no conflicts of interest in this work.
References
- WolfgangCLHermanJMLaheruDAKleinAPErdekMAFishmanEKHrubanRHRecent progress in pancreatic cancerCA Cancer J Clin201363531834823856911
- FogelELShahdaSSandrasegaranKA multidisciplinary approach to pancreas cancer in 2016: a reviewAm J Gastroenterol2017112453755428139655
- VincentAHermanJSchulickRHrubanRHGogginsMPancreatic cancerLancet2011378979160762021620466
- TorreLABrayFSiegelRLFerlayJLortet-TieulentJJemalAGlobal cancer statistics, 2012CA Cancer J Clin20156528710825651787
- ChenWZhengRBaadePDCancer statistics in China, 2015CA Cancer J Clin201666211513226808342
- ChenWSunKZhengRCancer incidence and mortality in China, 2014Chin J Cancer Res201830111229545714
- KandaMMatthaeiHWuJPresence of somatic mutations in most early-stage pancreatic intraepithelial neoplasiaGastroenterology20121427334730733.e9
- MaitraAAdsayNVArganiPMulticomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarrayMod Pathol200316990291213679454
- SeufferleinTBachetJBvan CutsemERougierPon behalf of the ESMO Guidelines Working Group, Group EGWPancreatic adenocarcinoma: ESMO-ESDO Clinical Practice Guidelines for diagnosis, treatment and follow-upAnn Oncol201223Suppl 7vii33vii4022997452
- SiegelRMaJZouZJemalACancer statistics, 2014CA Cancer J Clin201464192924399786
- GonzalezMAPinderSECallagyGMinichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancerJ Clin Oncol200321234306431314645419
- RamnathNHernandezFJTanD-FMCM2 is an independent predictor of survival in patients with non–small-cell lung cancerJ Clin Oncol200119224259426611709570
- DudderidgeTJKellyJDWollenschlaegerADiagnosis of prostate cancer by detection of minichromosome maintenance 5 protein in urine sedimentsBr J Cancer2010103570170720648010
- WilliamsGHSwinnRPrevostATDiagnosis of oesophageal cancer by detection of minichromosome maintenance 5 protein in gastric aspiratesBr J Cancer200491471471915266314
- Consortium GTThe genotype-tissue expression (GTEx) projectNat Genet201345658058523715323
- FerreiraPGMuñoz-AguirreMReverterFThe effects of death and post-mortem cold ischemia on human tissue transcriptomesNat Commun20189149029440659
- GTEx ConsortiumHuman genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humansScience2015348623564866025954001
- TangZLiCKangBGaoGLiCZhangZGEPIA: a web server for cancer and normal gene expression profiling and interactive analysesNucleic Acids Res201745W1W98W10228407145
- ShaulYDYuanBThiruPMERAV: a tool for comparing gene expression across human tissues and cell typesNucleic Acids Res201644D1D560D56626626150
- Cancer Genome Atlas Research NetworkGenomic Characterization of Pancreatic Ductal AdenocarcinomaCancer Cell201732218520328810144
- AndersSHuberWDifferential expression analysis for sequence count dataGenome Biol20101110R10620979621
- LiaoXHuangKHuangRGenome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomyOnco Targets Ther2017104493450628979141
- SubramanianATamayoPMoothaVKGene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profilesProc Natl Acad Sci U S A200510243155451555016199517
- MoothaVKLindgrenCMErikssonK-FPGC-1α-responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetesNat Genet200334326727312808457
- LiberzonABirgerCThorvaldsdóttirHGhandiMMesirovJPTamayoPThe Molecular Signatures Database hallmark gene set collectionCell Syst20151641742526771021
- BenjaminiYHochbergYControlling the false discovery rate: a practical and powerful approach to multiple testingJ Royal Stat Soc1995571289300
- ReinerAYekutieliDBenjaminiYIdentifying differentially expressed genes using false discovery rate controlling proceduresBioinformatics200319336837512584122
- BenjaminiYDraiDElmerGKafkafiNGolaniIControlling the false discovery rate in behavior genetics researchBehav Brain Res20011251–227928411682119
- MostafaviSRayDWarde-FarleyDGrouiosCMorrisQGeneMA-NIA: a real-time multiple association network integration algorithm for predicting gene functionGenome Biol20089Suppl 1S4
- MeringCvvon MeringCHuynenMJaeggiDSchmidtSBorkPSnelBSTRING: a database of predicted functional associations between proteinsNucleic Acids Res200331125826112519996
- KunnevDFreelandAQinMLeachRWWangJShenoyRMPruittSCEffect of minichromosome maintenance protein 2 deficiency on the locations of DNA replication originsGenome Res201525455856925762552
- ChongJPJThömmesPBlowJJThe role of MCM/P1 proteins in the licensing of DNA replicationTrends Biochem Sci19962131021068882583
- SuTTFollettePJO’FarrellPHQualifying for the license to replicateCell19958168258287781058
- StillmanBCell cycle control of DNA replicationScience19962745293165916638939847
- KearseySELabibKMCM proteins: evolution, properties, and role in DNA replicationBiochim Biophys Acta1998139821131369689912
- WoodwardAMGöhlerTLucianiMGExcess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stressJ Cell Biol2006173567368316754955
- BlowJJDuttaAPreventing re-replication of chromosomal DNANat Rev Mol Cell Biol20056647648615928711
- PruittSCBaileyKJFreelandAReduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancerStem Cells200725123121313217717065
- ChuangC-HWallaceMDAbratteCSouthardTSchimentiJCIncremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stressPLoS Genet201069e100111020838603
- ShimaNAlcarazALiachkoIA viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in miceNat Genet2007391939817143284
- WatanabeEOharaRIshimiYEffect of an MCM4 mutation that causes tumours in mouse on human MCM4/6/7 complex formationJ Biochem2012152219119822668557
- LeiMThe MCM complex: its role in DNA replication and implications for cancer therapyCurr Cancer Drug Targets20055536538016101384
- SaydamOSenolOSchaaij-VisserTBMComparative protein profiling reveals minichromosome maintenance (MCM) proteins as novel potential tumor markers for meningiomasJ Proteome Res20109148549419877719
- MukherjeeGMuralidharBBafnaUDLaskeyRAColemanNMCM immunocytochemistry as a first line cervical screening test in developing countries: a prospective cohort study in a regional cancer centre in IndiaBr J Cancer20079671107111117342084
- Saeb-ParsyKWilsonAScarpiniCDiagnosis of bladder cancer by immunocytochemical detection of minichromosome maintenance protein-2 in cells retrieved from urineBr J Cancer201210781384139122968648
- DaviesRJFreemanAMorrisLSAnalysis of minichromosome maintenance proteins as a novel method for detection of colorectal cancer in stoolLancet200235993211917191912057556
- WangYLiYZhangWYmRNA expression of minichromosome maintenance 2 in colonic adenoma and adenocarcinomaEur J Cancer Prev2009181404519077563
- AyaruLStoeberKWebsterGJDiagnosis of pancreaticobiliary malignancy by detection of minichromosome maintenance protein 5 in bile aspiratesBr J Cancer20089891548155418414413
- KeaneMGHuggettMTChapmanMHDiagnosis of pancreaticobiliary malignancy by detection of minichromosome maintenance protein 5 in biliary brush cytologyBr J Cancer2017116334935528081547
- ZhengTChenMHanSPlasma minichromosome maintenance complex component 6 is a novel biomarker for hepatocellular carcinoma patientsHepatology Res2014441313471356
- RezazadehFEbrahimiRAndisheh-TadbirAAshrafMJKhademiBEvaluation of the Ki-67 and MCM3 expression in cytologic smear of oral squamous cell carcinomaJ Dent2017183207211
- LeeYSHaS-AKimHJMinichromosome maintenance protein 3 is a candidate proliferation marker in papillary thyroid carcinomaExp Mol Pathol201088113814219818763
- AshkavandiZJNajvaniADTadbirAAPardisSRanjbarMAAshrafMJMCM3 as a novel diagnostic marker in benign and malignant salivary gland tumorsAsian Pac J Cancer Prev20131463479348223886132
- GambichlerTBreiningerARotterdamSAltmeyerPStückerMKreuterAExpression of minichromosome maintenance proteins in Merkel cell carcinomaJ Eur Acad Dermatol Venereol200923101184118819453809
- LiuHTakeuchiSMoroiYExpression of minichromosome maintenance 5 protein in proliferative and malignant skin diseasesInt J Dermatol200746111171117617988337
- YangJYLiDZhangYGuanBXGaoPZhouXCZhouCJThe expression of MCM7 is a useful biomarker in the early diagnostic of gastric cancerPathol Oncol Res201724236737228540486
- ChoyBLalondeAQueJWuTZhouZMCM4 and MCM7, potential novel proliferation markers, significantly correlated with Ki-67, Bmi1, and cyclin E expression in esophageal adenocarcinoma, squamous cell carcinoma, and precancerous lesionsHum Pathol20165712613527476776
- KimuraFOkayasuIKakinumaHSatohYKuwaoSSaegusaMWatanabeJDifferential diagnosis of reactive mesothelial cells and malignant mesothelioma cells using the cell proliferation markers minichromosome maintenance protein 7, geminin, topoisomerase II alpha and Ki-67Acta Cytol201357438439023860238
- KimuraFKawamuraJWatanabeJKamoshidaSKawaiKOkayasuIKuwaoSSignificance of cell proliferation markers (minichromosome maintenance protein 7, topoisomerase IIalpha and Ki-67) in cavital fluid cytology: can we differentiate reactive mesothelial cells from malignant cells?Diagn Cytopathol201038316116719821496
- RenBYuGTsengGCMCM7 amplification and overexpression are associated with prostate cancer progressionOncogene20062571090109816247466
- FengCJHjLJnLYjLLiaoGQExpression of Mcm7 and Cdc6 in oral squamous cell carcinoma and precancerous lesionsAnticancer Res2008286A3763376919189662
- DasMPrasadSBYadavSSOver expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesisPLoS One201387e6960723874974
- SunMWuGLiYExpression profile reveals novel prognostic biomarkers in hepatocellular carcinomaFront Biosci20102829840
- ZhongHChenBNevesHExpression of minichromosome maintenance genes in renal cell carcinomaCancer Manag Res2017963764729180899
- KangWTongJHChanAWChengASYuJToKMCM7 serves as a prognostic marker in diffuse-type gastric adenocarcinoma and siRNA-mediated knockdown suppresses its oncogenic functionOncol Rep20143152071207824647462
- KorkolopoulouPGivalosNSaettaAMinichromosome maintenance proteins 2 and 5 expression in muscle-invasive urothelial cancer: a multivariate survival study including proliferation markers and cell cycle regulatorsHum Pathol200536889990716112007
- GakiopoulouHKorkolopoulouPLevidouGMinichromosome maintenance proteins 2 and 5 in non-benign epithelial ovarian tumours: relationship with cell cycle regulators and prognostic implicationsBr J Cancer20079781124113417940502
- YangJRamnathNMoysichKBPrognostic significance of MCM2, Ki-67 and gelsolin in non-small cell lung cancerBMC Cancer20066120316882345
- YangCWenYLiHOverexpression of minichromosome maintenance 2 predicts poor prognosis in patients with gastric cancerOncol Rep201227113514221947329
- CzyzewskaJGuziαska-UstymowiczKPryczyniczAKemonaABandurskiRImmunohistochemical evaluation of Ki-67, PCNA and MCM2 proteins proliferation index (PI) in advanced gastric cancerFolia Histochem Cytobiol200947228929619995716
- LiuMJsLTianDPHuangBRosqvistSSuMMCM2 expression levels predict diagnosis and prognosis in gastric cardiac cancerHistol Histopathol201328448149223329420
- BurgerMDenzingerSHartmannAWielandW-FStoehrRObermannECMcm2 predicts recurrence hazard in stage Ta/T1 bladder cancer more accurately than CK20, Ki67 and histological gradeBr J Cancer200796111711171517505513
- LiuY-ZWangB-SJiangY-YMCMs expression in lung cancer: implication of prognostic significanceJ Cancer20178183641364729151950
- WangDLiQLiYWangHThe role of MCM5 expression in cervical cancer: correlation with progression and prognosisBiomed Pharmacother20189816517229253764
- VigourouxCCasseJ-MBattaglia-HsuS-FMethyl(R217)HuR and MCM6 are inversely correlated and are prognostic markers in non small cell lung carcinomaLung Cancer201589218919626013954
- HelfensteinAFrahmSOKramsMDrescherWParwareschRHassenpflugJMinichromosome maintenance protein (MCM6) in low-grade chondrosarcoma: distinction from enchondroma and identification of progressive tumorsAm J Clin Pathol2004122691291815539383
- SchraderCJanssenDKlapperWMinichromosome maintenance protein 6, a proliferation marker superior to Ki-67 and independent predictor of survival in patients with mantle cell lymphomaBr J Cancer200593893994516189522
- HottonJAgopiantzMLerouxAMinichromosome maintenance complex component 6 (MCM6) expression correlates with histological grade and survival in endometrioid endometrial adenocarcinomaVirchows Arch2018472462363329243125
- NishiharaKShomoriKFujiokaSMinichromosome maintenance protein 7 in colorectal cancer: implication of prognostic significanceInt J Oncol200833224525118636144
- ZhouY-MZhangX-FCaoLLiBSuiCJLiYMYinZFMCM7 expression predicts post-operative prognosis for hepatocellular carcinomaLiver Int201232101505150922784096
- ToyokawaGMasudaKDaigoYMinichromosome maintenance protein 7 is a potential therapeutic target in human cancer and a novel prognostic marker of non-small cell lung cancerMol Cancer20111016521619671
- FujiokaSShomoriKNishiharaKExpression of minichromosome maintenance 7 (MCM7) in small lung adenocarcinomas (pT1): prognostic implicationLung Cancer200965222322919144445
- MarneridesAVassilakopoulosTPBoltetsouEImmunohistochemical expression and prognostic significance of CCND3, MCM2 and MCM7 in Hodgkin lymphomaAnticancer Res201131103585359421965782
- TamuraTShomoriKHarukiTMinichromosome maintenance-7 and geminin are reliable prognostic markers in patients with oral squamous cell carcinoma: immunohistochemical studyJ Oral Pathol Med201039432833420136698
- ZhongXChenXGuanXOverexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosisHistopathology201566219220024805087
- IshibashiYKinugasaTAkagiYMinichromosome maintenance protein 7 is a risk factor for recurrence in patients with Dukes C colorectal cancerAnticancer Res20143484569457525075101
- WintherTLTorpSHMCM7 expression is a promising predictor of recurrence in patients surgically resected for meningiomasJ Neurooncol2017131357558327868157
- FristrupNBirkenkamp-DemtröderKReinertTMulticenter validation of Cyclin D1, MCM7, TRIM29, and UBE2C as prognostic protein markers in non-muscle–invasive bladder cancerAm J Pathol2013182233934923201130
- ColiAAsaSLFaddaGMinichromosome maintenance protein 7 as prognostic marker of tumor aggressiveness in pituitary adenoma patientsEur J Endocrinol2016174330731426620390
- OtaTClaytonACMinotDMShridharVHartmannLCGilksCBChienJRMinichromosome maintenance protein 7 as a potential prognostic factor for progression-free survival in high-grade serous carcinomas of the ovaryMod Pathol201124227728721076460
- PengY-PZhuYYinL-DThe expression and prognostic roles of MCMs in pancreatic cancerPLoS One20161110e016415027695057
- Komamura-KohnoYKarasawa-ShimizuKSaitohTSatoMHanaokaFTanakaSIshimiYSite-specific phosphorylation of MCM4 during the cell cycle in mammalian cellsFEBS J200627361224123916519687
- FujitaMYamadaCTsurumiTHanaokaFMatsuzawaKInagakiMCell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinaseJ Biol Chem19982732717095171019642275
- IshimiYKomamura-KohnoYKarasawa-ShimizuKYamadaKLevels of MCM4 phosphorylation and DNA synthesis in DNA replication block checkpoint controlJ Struct Biol20041461–223424115037254
- IshimiYKomamura-KohnoYKwonH-JYamadaKNakanishiMIdentification of MCM4 as a target of the DNA replication block checkpoint systemJ Biol Chem200327827246442465012714602
- CaseyJPNobbsMMcgettiganPLynchSEnnisSRecessive mutations in MCM4 / PRKDC cause a novel syndrome involving a primary immunodeficiency and a disorder of DNA repairJ Med Genet201249424224522499342
- YunHJHyunSKParkJHKimBWKwonHJWiddrol activates DNA damage checkpoint through the signaling Chk2–p53–Cdc25A–p21–MCM4 pathway in HT29 cellsMol Cell Biochem20123631–228128922160829
