Abstract
Background
The death-associated protein kinase (DAPK) gene is an important member of the apoptotic pathway and is inactivated by abnormal methylation in numerous cancers, including nasopharyngeal carcinoma (NPC). However, the diagnostic value of DAPK methylation in brushing samples and tissue samples of NPC remains unclear.
Methods
We conducted a systematic meta-analysis based on 17 studies (including 386 tissue cases, 233 brushing cases, and 296 blood cases).
Results
Our results revealed an association between methylated DAPK and increased risk of NPC in blood, brushing, and tissue samples. In addition, the comparison of the pooled sensitivity, specificity, and area under the curve of methylated DAPK in brushing and tissue samples demonstrated the non-inferior effectiveness of methylated DAPK in brushing samples to monitor the development of NPC.
Introduction
Nasopharyngeal carcinoma (NPC) is a cancer arising from the epithelial cells lining the nasopharynx. It has a wide geographic and racial distribution worldwide. The occurrence of NPC is rare in most parts of the world, but not in People’s Republic of China. It is endemic in southern People’s Republic of China, including Hong Kong, with a reported annual incidence of up to 50 cases per 100,000 people.Citation1 Approximately 60,000 new NPC cases and 34,100 deaths from NPC were projected to occur in 2015 in People’s Republic of China.Citation2
As in other major human cancers, the progression of NPC is a multistep process involving interactions between multiple factors, including Epstein-Barr virus (EBV) infection,Citation3,Citation4 consumption of salted food,Citation5–Citation7 cigarette smoking,Citation8,Citation9 and alcohol consumption.Citation10,Citation11 Among these, EBV infection is necessary for NPC progression. By adulthood, approximately 90% of individuals are EBV infected.Citation12,Citation13 Although EBV is a ubiquitous pathogen, EBV-associated NPC develops in only a small fraction of infected individuals. Thus, it is believed that genetic factors may contribute significantly to the high risk of NPC in this population. To determine the differences present in the subset of EBV-positive individuals who develop cancer, multiple genome-wide studies have examined genetic and epigenetic abnormalities in specific oncogenes and tumor suppressor genes (TSGs).Citation14–Citation16 In recent years, promoter methylation has been recognized as a common mechanism of inactivating TSGs in the tumorigenesis of NPC.Citation17,Citation18 Because DNA hypermethylation is one of the earliest molecular alterations during malignant transformation in human epithelial cells and often occurs earlier than the morphological abnormalities of cancers,Citation19,Citation20 the analysis of promoter methylation of TSGs may be a promising method for the detection of NPC.
Death-associated protein kinase (DAPK) is encoded by the DAPK gene, which is a novel serine/threonine kinase required for interferon gamma-induced apoptotic cell death.Citation21 Numerous cancer cell clones with highly aggressive metastatic behavior lack DAP kinase expression, whereas the clones with low metastatic capabilities express the protein.Citation22,Citation23 Restoration of DAP kinase in highly metastatic cancer cells can suppress the metastatic ability of these cancer cells.Citation22 As a novel TSG, the expression of DAP kinase is repressed in several types of human cancers by hypermethylation in the promoter CpG region of the gene,Citation24,Citation25 including in NPC.Citation26,Citation27 However, the diagnostic power of DAPK methylation in NPC has not been investigated.
In the current study, we performed a meta-analysis of 17 studies to assess the association of DAPK methylation with the risk of NPC and implemented a diagnostic meta-analysis to evaluate the diagnostic potential of DAPK methylation for NPC.
Materials and methods
Literature search strategy
We performed a comprehensive literature search from a range of electronic databases, including PubMed, Embase, Google Scholar, and Web of Science (last search updated in January 2018) without language restrictions. The following search keywords were used: (“methylation” or “DNA methylation” or “promoter methylation” or “demethylation” or “hypermethylation”) and (“nasopharyngeal cancer” or “nasopharyngeal neoplasm” or “nasopharyngeal carcinoma” or “NPC”) and (“DAPK” or “death associated protein kinase”).
Selection criteria
The following predefined criteria were used to evaluate the eligibility of included studies: 1) the study design must be a case–control study focused on the association between DAPK promoter methylation and NPC patients and 2) the study must provide sufficient information about DAPK promoter methylation to calculate odds ratio (OR) and 95% CI. The study was excluded if it did not meet the inclusion criteria. If the authors had published multiple studies using the same population, only the most recent or the largest-sample-size publication was used in our meta-analysis.
Data extraction
All the relevant data of the eligible studies were retrieved independently by all the authors of this study. The following information was extracted: the first author’s name, the published year, the race distribution of the study subjects, the source of the samples, the number of participants, and the frequency of DAPK methylation.
Statistical analysis
The strength of the association between methylated DAPK and the risk of NPC is represented by the pooled overall OR across all the eligible studies. The heterogeneity of all eligible studies was quantified with the I2 statistic and χ2 test with the corresponding P-value.Citation28 A DerSimonian–Laird (D+L) model was applied to calculate pooled ORs when there existed heterogeneity in the meta-analysis (I2 > 50%, χ2 test with P < 0.05). Otherwise, a Mantel–Haenszel (M−H) model was used.Citation28 A meta-regression was performed to identify the source of heterogeneity. Sensitivity analysis was performed to assess the stability of our results by omitting single studies in the meta-analysis iteration to determine the effect of the individual data on the overall pooled OR. The stability of our results was tested by switching between the D+L and M−H models. Publication bias was quantitatively estimated by Egger’s linear regression test.
For the diagnostic meta-analysis, pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and their corresponding 95% CIs were calculated. The PLR is calculated as sensitivity/(1–specificity), and the NLR is calculated as (1–sensitivity)/specificity. The DOR is a measure that combines sensitivity and specificity and is calculated as PLR/NLR.Citation29 The Fagan plots, assessing the clinical utility of a tested indicator, were drawn based on the values of PLR and NLR. We evaluated pre-test probabilities of 25% and 50% versus corresponding post-test probabilities.Citation30 Summary receiver operation characteristic curves (SROCs) with the area under the receiver operating characteristic curve (AUC) were generated. All the data analyses were accomplished by STATA-12.0 software (Stata Corporation, College Station, TX, USA). All P-values were two sided, and a P-value less than 0.05 was deemed significant.
Results
Study characteristics
First, 311 articles were collected by electronic searches in PubMed, Embase, Google Scholar, and Chinese National Knowledge Infrastructure. The selection process of eligible studies is illustrated in . After carefully filtering all the potential papers according to the selection criteria, a total of 17 studies were included in the meta-analysis. Among them, three studies evaluated the association between DAPK methylation and the risk of NPC using blood samples,Citation31–Citation33 and five studies used brushing samples.Citation33–Citation36 Nine other studies assessed the correlation of DAPK methylation and the risk of NPC using NPC and non-tumorous tissue samples.Citation17,Citation26,Citation27,Citation35–Citation40 shows the main characteristics of the included studies.
Table 1 General characteristics of studies included in the current meta-analysis
Association of DAPK methylation in NPC and controls
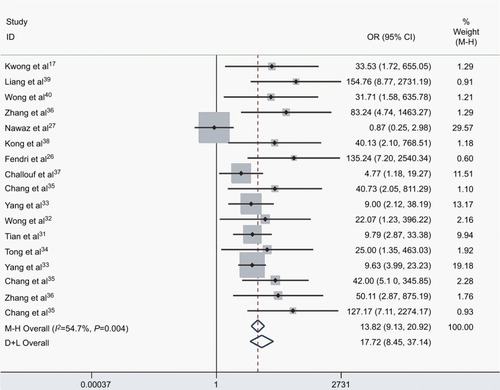
We assessed the difference in DAPK methylation between NPC and normal controls in the 17 studies, which included 915 NPC patients and 404 controls. As there was heterogeneity across studies (I2 = 54.7%, P = 0.004), we changed the fixed-effects model (M−H) and random-effects model (D+L) to affirm the reliability of our results (OR(M−H) = 13.82, 95% CI: [9.13, 20.92]; OR(D+L) = 17.72, 95% CI: [8.45, 37.14]). Besides, sensitivity analysis was also performed to support the robustness of our results (). The pooled ORs strongly suggested that DAPK methylation was associated with increased risk of NPC (). Egger’s test revealed no evidence of publication bias (P = 0.58; ).
Table 2 Sensitivity analysis of pooled OR for DAPK methylation between NPC and controls
Figure 2 The forest plot for the association between DAPK methylation and the risk of NPC by the fixed-effects model (M−H) and random-effects model (D+L) in NPC vs controls.
Abbreviations: NPC, nasopharyngeal carcinoma; M−H, Mantel–Haenszel; D+L, DerSimonian–Laird; OR, odds ratio; CI, confidence interval.
Subgroup analysis of DAPK methylation in NPC and controls
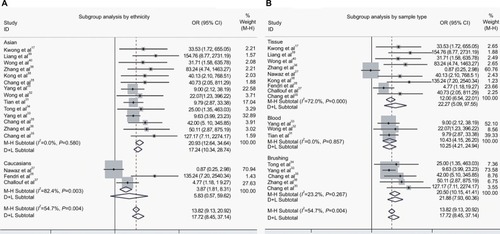
To ascertain the source of heterogeneity, a meta-regression was performed. Our results attributed the heterogeneity to the three studies on Caucasians (). Therefore, subgroup analyses by ethnicity were conducted. As seen in , a significant decrease was observed in heterogeneity after exclusion of the three studies on Caucasians (I2 = 0.0%, P = 0.58, ). Additionally, the pooled OR illustrated the association of DAPK methylation and elevated risk of NPC (OR(M−H) = 20.93, 95% CI: [12.64, 34.64]).
Table 3 Meta-regression analysis of DAPK methylation in NPC vs controls
Figure 4 The forest plots for the association of DAPK methylation and the risk of NPC by subgroup analyses.
Notes: (A) Subgroup analysis by ethnicity; (B) Subgroup analysis by sample type.
Abbreviations: NPC, nasopharyngeal carcinoma; OR, odds ratio; CI, confidence interval; M−H, Mantel-Haenszel ; D+L, DerSimonian–Laird.
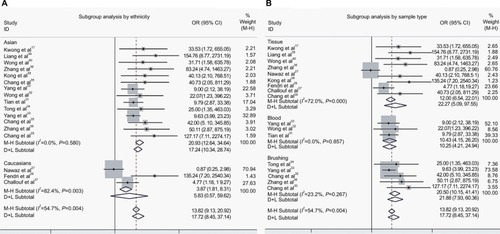
In addition, we performed a subgroup analysis by sample type, and the results showed that the pooled OR was 22.27 for tissue samples, 10.43 for blood samples, and 20.50 for brushing samples (tissue: OR(D+L) = 22.27, 95% CI [5.09, 97.55]; blood: OR(M−H) = 10.43, 95% CI [4.15, 26.20]; brushing: OR(M−H) = 20.50, 95% CI [10.15, 41.41]; ).
Diagnostic value of DAPK methylation for NPC and controls
As shown in , the pooled OR in tissue samples was congruent with that in brushing samples, implying that the DAPK methylation in brushing samples may serve as a useful and noninvasive biomarker for NPC. However, the comparison of the diagnostic capability of DAPK methylation for NPC in tissue samples and in brushing sample has not been investigated. Therefore, we performed a diagnostic meta-analysis of 9 studies on NPC tissue samples and a separate one of 5 studies on NPC brushing samples. The summary specificity and sensitivity of methylated DAPK for distinguishing NPC from control tissue samples were 0.99 and 0.69 (0.55–0.80), respectively (tissue: specificity = 0.99, 95% CI [0.85, 1.00]; sensitivity = 0.69, 95% CI [0.55, 0.80]; ). The summary specificity and sensitivity of methylated DAPK for identification of NPC from control brushing samples were 0.98 and 0.58, respectively (brushing: specificity = 0. 98, 95% CI [0.85, 1.00]; sensitivity = 0. 58, 95% CI [0.50, 0.67]; ).
Figure 5: Forest sensitivity and specificity of methylated DAPK for diagnosis of NPC.
Notes: (A) Forest sensitivity and specificity of methylated DAPK for diagnosis of NPC using tissue sample; (B) Forest sensitivity and specificity of methylated DAPK for diagnosis of NPC using brushing sample.
Abbreviation: NPC, nasopharyngeal carcinoma; OR, odds ration; CI, confidence interval.
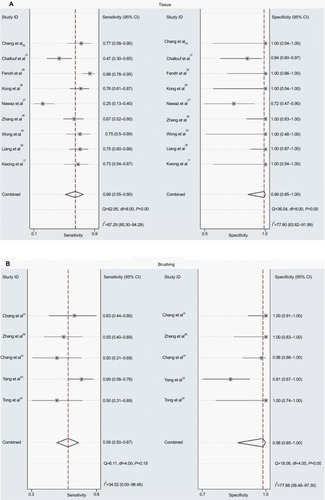
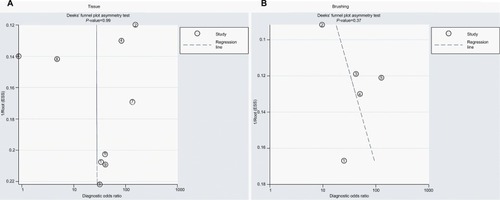
The SROCs based on specificity and sensitivity are shown in . The AUC was 0.92 for tissue and 0.71 for brushing samples (tissue: AUC = 0. 92, 95% CI [0.90–0.94], ; brushing: AUC = 0.71, 95% CI [0.67, 0.75], ). In addition, the summary DOR, another diagnostic-strength parameter that indicates better diagnostic strength with higher values, was 184 for tissue and 87 for brushing (tissue: DOR = 184, 95% CI [9, 3725]; brushing: DOR = 87, 95% CI [9, 865]). There was no publication bias in this diagnostic meta-analysis ().
Figure 6 SROC plots with best fitting asymmetric curve of methylated DAPK for NPC by sample type.
Notes: (A) SROC plots with best fitting asymmetric curve of methylated DAPK for NPC by tissue sample; (B) SROC plots with best fitting asymmetric curve of methylated DAPK for NPC by brushing sample.
Abbreviations: SENS, sensitivity; SPEC, specificity; SROC, summary of receiver-operator characteristic; NPC, nasopharyngeal carcinoma; AUC, area under the receiver operating characteristic curve.
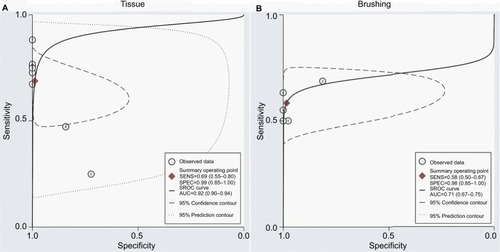
Figure 7 The publication bias of methylated DAPK for diagnosis of NPC by sample type.
Notes: (A) The publication bias of methylated DAPK for diagnosis of NPC by tissue sample. (B) The publication bias of methylated DAPK for diagnosis of NPC by brushing sample.
Abbreviations: ESS, effective sample size; NPC, nasopharyngeal carcinoma.
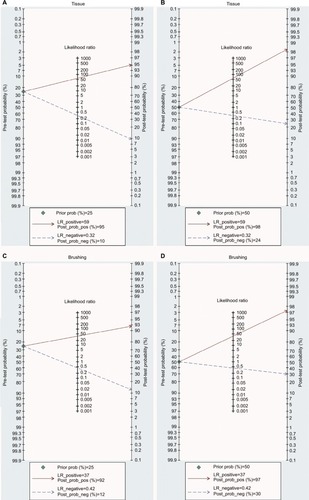
The abovementioned results confirm that the detection of DAPK methylation could serve as an auxiliary technology for the diagnosis of NPC. Therefore, it is necessary to evaluate the clinical value of DAPK methylation during clinical practice. The PLR and NLR are effective indicators of clinical utility, as is the Fagan plot. The PLR and NLR for tissue were 58.8 and 0.32, respectively (tissue: PLR = 58.8, 95% CI [3.7, 930.0]; NLR = 0.32, 95% CI [0.21, 0.47]), while the PLR and NLR for brushing samples were 36.7 and 0.42, respectively (brushing: PLR = 36.7, 95% CI [3.7, 367.3]; NLR = 0.42, 95% CI [0.35, 0.51]). As indicated by PLR in tissue, NPC patients had a nearly a 59 times higher chance of positive detection of DAPK methylation than in control tissue samples. The NLR indicated that normal control tissue samples had a threefold greater chance (the reciprocal of the value of NLR) of having unmethylated DAPK than NPC patients. The PLR and NLR for brushing samples indicated a nearly 37 times higher chance of positive detection of DAPK methylation in NPC brushing samples than in controls and twofold greater chance of having unmethylated DAPK in control brushing samples than in NPC patients.
The Fagan plot was generated for the visual presentation of the diagnostic performance of the detection of methylated DAPK. As shown in , when the prior probability was taken as 25% and 50%, the Fagan plot illustrated that the probability of an individual being diagnosed with NPC was 95% and 98%, respectively, following a methylated DAPK in tissue samples. However, the probability of an exclusion diagnosis of NPC was 16% and 26% following a non-methylated DAPK in tissue samples. As illustrated in , with prior probabilities of 25% and 50%, the Fagan plot illustrated that the probability of an individual being diagnosed with NPC was 93% and 97%, respectively, following a methylated DAPK in brushing samples. However, the probability of an exclusion diagnosis of NPC was 13% and 20% following a non-methylated DAPK in brushing samples. The results of our diagnostic meta-analysis imply that the detection of methylated DAPK in brushing samples could serve as an effective biomarker for diagnosis of NPC.
Figure 8 Fagan plots analysis to illustrate the clinical utility of methylated DAPK for identification of NPC by sample type.
Notes: (A) The post-test probability of NPC was 95% at a pretest probability of 25% by tissue samples. (B) The post-test probability of NPC was 98% at a pretest probability of 50% by tissue samples. (C) The post-test probability of NPC was 92% at a pretest probability of 25% by brushing sample. (D) The post-test probability of NPC was 97% at a pretest probability of 50% by brushing sample.
Discussion
The incidence of NPC is rare in western populations, with rates below 2 per 100,000 person-years.Citation41 NPC is much more common in People’s Republic of China and in the Arctic region, with incidence rates up to 30 per 100,000 persons.Citation41 However, even within Asian populations, dramatic differences in NPC incidence are observed across regions. Higher incidences of NPC are found in urban areas than in rural areas.Citation42
Although multiple specific environmental factors, including early exposure to salted food and latent EBV infection, have been suggested to be risk factors in the endemic regions, the predisposition to NPC among southern Chinese population strongly suggests the involvement of both genetic and epigenetic susceptibility and environmental factors.Citation43,Citation44 Genome-wide linkage analyses of high-risk Chinese NPC identified several candidate NPC susceptibility loci, including chromosome 3p21.Citation44,Citation45 Many TSGs, such as RASSF1A and MLH1, have been isolated from this region.Citation46,Citation47 Inactivation of multiple TSGs attributed to high frequencies of deletion of this region has been associated with the progression of NPC.Citation14,Citation48 Additionally, aberrant methylation of the 5′ CpG island is also a major mechanism for the silencing of these genes.Citation49–Citation51
DNA methylation is the process of formation of methylcytosine in DNA by the addition of a methyl group to a cytosine.Citation52 Neoplastic cells simultaneously harbor widespread genomic hypomethylation and more regional areas of hypermethylation. Each component of methylation imbalance may contribute to tumor progression.Citation53,Citation54 Hypermethylation of gene promoter regions is associated with gene repression and can be considered an alternative modification to coding mutations that induces the inactivation of TSGs (such as P16INK4a, MGMT, GSTP1, and APC) in numerous human cancers.Citation55–Citation58 The dysregulation of TSGs resulting in the imbalance of biological process and uncontrolled cell proliferation is contributed to the transformation of neoplastic cell.Citation59 Among these abnormal methylated genes in human cancers, the hypermethylation of CDKN2A is well characterized that is involved in tumorigenesis by inducing the loss of negative regulator of cell proliferation.Citation60 Besides, the identities of the hypermethylated regions can vary between cancer types.Citation61 The use of the hypermethylation events of MGMT in glioma and GSTP1 in prostate cancerCitation62 is effective for diagnosis of cancer, indicating that alterations of epigenetic markers could be used as cancer biomarkers.
DAPK, encoded by the DAPK gene, belongs to the DAPK family. The DAPK family contains three closely related serine/threonine kinases, named DAPK, ZIPK, and DRP-1.Citation21 Several lines of evidence indicate that the most studied member of the DAPK family, DAPK, has tumor and metastasis suppressor properties.Citation26,Citation63 DAPK downregulation or inactivation through epigenetic modification, especially DNA methylation, has been observed in a number of metastatic cancers.Citation64,Citation65 The imbalance of proliferation and apoptosis, partly induced by the inactivation of the apoptotic pathway, has been considered as one of the hallmarks of cancer, specifically the initiation and progression of human cancers, including NPC.Citation66 The dysfunction of apoptosis-related genes could decrease apoptosis induced by chemotherapy. Thus, the inactivation of the apoptotic pathway by aberrant methylation has been associated with chemoresistance.Citation67,Citation68 As an important member of the apoptotic pathway, the role of abnormal hypermethylation of DAPK in NPC has been investigated by many researchers.Citation31–Citation36 The detection of DAPK methylation in different NPC patients may be one reason for the inconsistent conclusions about the association between DAPK methylation and the risk of NPC.Citation33,Citation35 To solve this problem, we performed a meta-analysis and conducted a subgroup analysis by sample source, and our results support the correlation between hypermethylated DAPK and increased risk of NPC, which was also confirmed by subgroup analysis of sample type. Moreover, the pooled OR in brushing samples was close to that in tissue samples, indicating that the detection of methylated DAPK in brushing samples could serve as an important alternative non-invasion measurement for diagnosis of NPC. Traditional diagnosis of NPC is made by biopsy of the nasopharyngeal mass. Fused positron emission tomography/computed tomography is a valuable imaging tool in patients for staging diagnosis of NPC. However, NPC is commonly diagnosed late due to its deep location and vague symptoms.Citation69 Thus, by measuring the nuclear DNA content, DNA diploidy was found to occur earlier in the progression from premalignant to malignant head and neck squamous cell carcinomas (including NPC). However, the diagnostic strength of methylated DAPK has not been investigated in NPC.
The current study aims to demonstrate that methylation of DAPK was readily applicable for routine diagnostic work. Therefore, diagnostic meta-analyses were performed in brushing samples and tissue samples separately to assess the power of methylated DAPK in distinguishing NPC from control tissue. Since the minimum number of included studies for a diagnostic meta-analysis is four, the diagnostic strength of methylated DAPK in blood samples was unable to be evaluated. The summary specificity and sensitivity of methylated DAPK for tissue samples were 0.99 and 0.69, respectively, and for brushing samples, they were 0.98 and 0.58, respectively, which shows the non-inferior effect for early monitoring of NPC in brushing samples. Fagan plots were drawn based on the values of PLR and NLR to assess the clinical utility of methylated DAPK. The Fagan plot is calculated based on Bayes’ rule, which is used to formalize how the pre-test probability of the risk of NPC is changed by the detection of methylated DAPK to yield the post-test probability of the risk of NPC.Citation30 From our results, in both diagnostic meta-analyses, the post-test probability of NPC risk increased to more than 90% when an individual with 25% of pre-test probability of NPC had a positive result of DAPK methylation. For the exclusion diagnosis, the post-test probability of NPC risk decreased to less than 30% when an individual with the 50% pre-test probability of NPC had a negative result of DAPK methylation. According to the results of this diagnostic meta-analysis, the detection of methylated DAPK in brushing samples for distinguishing NPC from non-tumor samples could serve as an alternative non-invasive biomarker.
In summary, this integrated analysis demonstrated the correlation of methylated DAPK and increased risk of NPC. In addition, the detection of methylated DAPK in brushing samples of NPC could serve as a promising alternative measurement for monitoring the initiation of NPC. Well-designed prospective studies with larger sample sizes will be indispensable to confirm our results.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (no. 81670920), Zhejiang Provincial Natural Science Foundation of China (nos. LY14H160003, LY15H130003), the Scientific Innovation Team Project of Ningbo (no. 2012B82019), Ningbo Social Developmental Key Research Project (no. 2012C5015), Medical and Health Research Project of Zhejiang Province (nos. 2014PYA017; 2012ZDA042; 2017KY133), Ningbo Health Branding Subject Fund (no. PPXK2018-02), and Ningbo Natural Science Foundation (nos. 2012A610208; 2017A610236, 2015A610221).
Disclosure
The authors report no conflicts of interest in this work.
References
- LuoJChiaKSChiaSEReillyMTanCSYeWSecular trends of nasopharyngeal carcinoma incidence in Singapore, Hong Kong and Los Angeles Chinese populations, 1973–1997Eur J Epidemiol200722851352117594525
- ChenWZhengRBaadePDCancer statistics in China, 2015CA Cancer J Clin201666211513226808342
- GhoshSKSinghASMondalRKapfoWKhamoVSinghYIDysfunction of mitochondria due to environmental carcinogens in nasopharyngeal carcinoma in the ethnic group of Northeast Indian populationTumour Biol20143576715672424711137
- TsaoSWYipYLTsangCMEtiological factors of nasopharyngeal carcinomaOral Oncol201450533033824630258
- BelbarakaRLalyaIBoulaamaneLTaziMBenjaafarNErrihaniHLes facteurs de risque alimentaires du carcinome indifférencié du nasopharynx: Une étude cas témoin. [Dietary risk factors of undifferenced nasopharyngeal carcinoma: a case-control study]Tunis Med2013916406409 French [with English abstract]23868040
- GallicchioLMatanoskiGTaoXGAdulthood consumption of preserved and nonpreserved vegetables and the risk of nasopharyngeal carcinoma: a systematic reviewInt J Cancer200611951125113516570274
- JiaWHLuoXYFengBJTraditional Cantonese diet and nasopharyngeal carcinoma risk: a large-scale case-control study in Guangdong, ChinaBMC Cancer20101044620727127
- FachirohJSangrajrangSJohanssonMTobacco consumption and genetic susceptibility to nasopharyngeal carcinoma (NPC) in ThailandCancer Causes Control201223121995200223085811
- XueWQQinHDRuanHLShugartYYJiaWHQuantitative association of tobacco smoking with the risk of nasopharyngeal carcinoma: a comprehensive meta-analysis of studies conducted between 1979 and 2011Am J Epidemiol2013178332533823785114
- ChenLGallicchioLBoyd-LindsleyKAlcohol consumption and the risk of nasopharyngeal carcinoma: a systematic reviewNutr Cancer200961111519116871
- RuanHLXuFHLiuWSAlcohol and tea consumption in relation to the risk of nasopharyngeal carcinoma in Guangdong, ChinaFront Med China20104444845621110141
- ChenCYHuangKYShenJHTsaoKCHuangYCA large-scale seroprevalence of Epstein-Barr virus in TaiwanPLoS One2015101e011583625615611
- DowdJBPalermoTBriteJMcDadeTWAielloASeroprevalence of Epstein-Barr virus infection in U.S. children ages 6–19, 2003–2010PLoS One201385e6492123717674
- HuiABLoKWLeungSFDetection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisationInt J Cancer199982449850310404061
- LoKWHuangDPGenetic and epigenetic changes in nasopharyngeal carcinomaSemin Cancer Biol200212645146212450731
- LoKWTeoPMHuiABHigh resolution allelotype of microdissected primary nasopharyngeal carcinomaCancer Res200060133348335310910036
- KwongJLoKWToKFTeoPMJohnsonPJHuangDPPromoter hypermethylation of multiple genes in nasopharyngeal carcinomaClin Cancer Res20028113113711801549
- RountreeMRBachmanKEHermanJGBaylinSBDNA methylation, chromatin inheritance, and cancerOncogene200120243156316511420732
- BrooksJDWeinsteinMLinXCG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasiaCancer Epidemiol Biomarkers Prev1998765315369641498
- LamyASesboüéRBourguignonJAberrant methylation of the CDKN2a/p16INK4a gene promoter region in preinvasive bronchial lesions: a prospective study in high-risk patients without invasive cancerInt J Cancer2002100218919312115568
- FeinsteinEDruckTKasturyKAssignment of DAP1 and DAPK—genes that positively mediate programmed cell death triggered by IFN-gamma—to chromosome regions 5p12.2 and 9q34.1, respectivelyGenomics19952913053078530096
- DansranjavinTMöbiusCTannapfelAE-cadherin and DAP kinase in pancreatic adenocarcinoma and corresponding lymph node metastasesOncol Rep20061551125113116596173
- InbalBCohenOPolak-CharconSDAP kinase links the control of apoptosis to metastasisNature199739066561801849367156
- KatzenellenbogenRABaylinSBHermanJGHypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignanciesBlood199993124347435310361133
- KissilJLFeinsteinECohenODAP-kinase loss of expression in various carcinoma and B-cell lymphoma cell lines: possible implications for role as tumor suppressor geneOncogene19971544034079242376
- FendriAMasmoudiAKhabirAInactivation of RASSF1A, RARbeta2 and DAP-kinase by promoter methylation correlates with lymph node metastasis in nasopharyngeal carcinomaCancer Biol Ther20098544445119221469
- NawazIMoumadKMartorelliDDetection of nasopharyngeal carcinoma in Morocco (North Africa) using a multiplex methylation-specific PCR biomarker assayClin Epigenetics201578926300994
- CooryMDComment on: Heterogeneity in meta-analysis should be expected and appropriately quantifiedInt J Epidemiol2010393932 author reply 93319349478
- JacksonDThe power of the standard test for the presence of heterogeneity in meta-analysisStat Med200625152688269916374903
- HellmichMLehmacherWA ruler for interpreting diagnostic test resultsMethods Inf Med200544112412615778803
- TianFYipSPKwongDLLinZYangZWuVWPromoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinomaCancer Epidemiol201337570871323790641
- WongTSKwongDLShamJSWeiWIKwongYLYuenAPQuantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinomaClin Cancer Res20041072401240615073117
- YangXDaiWKwongDLEpigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution meltingInt J Cancer20151364E127E13525196065
- TongJHTsangRKLoKWQuantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinomaClin Cancer Res2002882612261912171892
- ChangHWChanAKwongDLWeiWIShamJSYuenAPEvaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patientInt J Cancer2003105685185512767073
- ZhangZSunDHutajuluSHDevelopment of a non-invasive method, multiplex methylation specific PCR (MMSP), for early diagnosis of nasopharyngeal carcinomaPLoS One2012711e4590823144779
- ChalloufSZiadiSZaghdoudiRKsiaaFBen GacemRTrimecheMPatterns of aberrant DNA hypermethylation in nasopharyngeal carcinoma in Tunisian patientsClin Chim Acta20124137–879580222296674
- KongWJZhangSGuoCKEffect of methylation-associated silencing of the death-associated protein kinase gene on nasopharyngeal carcinomaAnticancer Drugs200617325125916520653
- LiangHLiLChenQLiXMethylation of DAPK gene promoter in nasopharyngeal carcinomaMod Oncol2015912131215 Chinese
- WongTSChangHWTangKCHigh frequency of promoter hypermethylation of the death-associated protein-kinase gene in nasopharyngeal carcinoma and its detection in the peripheral blood of patientsClin Cancer Res20028243343711839660
- FormanDBrayFBrewsterDHCancer Incidence in Five ContinentsXIARC Scientific Publication No. 164LyonInternational Agency for Research on Cancer2013 Available from: http://www.iarc.fr/en/publications/pdfs-online/epi/sp164/Accessed July 30, 2018
- WeiKRZhengRSZhangSWLiangZHLiZMChenWQNasopharyngeal carcinoma incidence and mortality in China, 2013Chin J Cancer20173619029122009
- FriborgJWohlfahrtJMelbyeMFamilial risk and clustering of nasopharyngeal carcinoma in Guangdong, ChinaCancer20051031211 author reply 211–21215540234
- XiongWZengZYXiaJHA susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinomaCancer Res20046461972197415026332
- FengBJHuangWShugartYYGenome-wide scan for familial nasopharyngeal carcinoma reveals evidence of linkage to chromosome 4Nat Genet200231439539912118254
- DammannRLiCYoonJHChinPLBatesSPfeiferGPEpigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3Nat Genet200025331531910888881
- HemminkiAPeltomäkiPMecklinJPLoss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancerNat Genet1994844054107894494
- LoKTsaoSLeungSChoiPLeeJHuangDDetailed deletion mapping on the short arm of chromosome-3 in nasopharyngeal carcinomasInt J Oncol1994461359136421567062
- CapelEFléjouJFHamelinRAssessment of MLH1 promoter methylation in relation to gene expression requires specific analysisOncogene200726547596760017546041
- MorrisseyCMartinezAZatykaMEpigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinomaCancer Res200161197277728111585766
- WagnerKJCooperWNGrundyRGFrequent RASSF1A tumour suppressor gene promoter methylation in Wilms’ tumour and colorectal cancerOncogene200221477277728212370819
- BaylinSBHermanJGGraffJRVertinoPMIssaJPAlterations in DNA methylation: a fundamental aspect of neoplasiaAdv Cancer Res1998721411969338076
- BaylinSBMakosMWuJJAbnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progressionCancer Cells19913103833901777359
- NassSJHermanJGGabrielsonEAberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancerCancer Res200060164346434810969774
- CalmonMFColomboJCarvalhoFMethylation profile of genes CDKN2A (p14 and p16), DAPK1, CDH1, and ADAM23 in head and neck cancerCancer Genet Cytogenet20071731313717284367
- GersonSLMGMT: its role in cancer aetiology and cancer therapeuticsNat Rev Cancer20044429630715057289
- MillarDSOwKKPaulCLRussellPJMolloyPLClarkSJDetailed methylation analysis of the glutathione S-transferase pi (GSTP1) gene in prostate cancerOncogene19991861313132410022813
- TsuchiyaTTamuraGSatoKDistinct methylation patterns of two APC gene promoters in normal and cancerous gastric epitheliaOncogene200019323642364610951570
- EvanGIVousdenKHProliferation, cell cycle and apoptosis in cancerNature200141134211357141
- TrzeciakLHennigEKolodziejskiJNowackiMOstrowskiJMutations, methylation and expression of CDKN2a/p16 gene in colorectal cancer and normal colonic mucosaCancer Lett20011631172311163104
- CostelloJFFruhwaldMCSmiragliaDJAberrant CpG-island methylation has non-random and tumour-type-specific patternsNat Genet200024213213810655057
- EstellerMCornPGUrenaJMGabrielsonEBaylinSBHermanJGInactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasiaCancer Res19985820451545189788592
- ChenHYLeeYRChenRHThe functions and regulations of DAPK in cancer metastasisApoptosis201419236437024166138
- FischerJROhnmachtURiegerNPromoter methylation of RASSF1A, RARbeta and DAPK predict poor prognosis of patients with malignant mesotheliomaLung Cancer200654110911616893590
- MittagFKuesterDViethMDAPK promotor methylation is an early event in colorectal carcinogenesisCancer Lett20062401697516246486
- HanahanDWeinbergRAThe hallmarks of cancerCell20001001577010647931
- IshiguroMIidaSUetakeHEffect of combined therapy with low-dose 5-aza-2′-deoxycytidine and irinotecan on colon cancer cell line HCT-15Ann Surg Oncol20071451752176217195906
- TeodoridisJMStrathdeeGBrownREpigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarkerDrug Resist Updat200474–526727815533764
- ChenYKSuCTDingHJClinical usefulness of fused PET/CT compared with PET alone or CT alone in nasopharyngeal carcinoma patientsAnticancer Res2006262B1471147716619560