Abstract
Background
Long intergenic non-protein coding RNA, a regulator of reprogramming (ROR), has been found to play an oncogene role in various human malignant tumors. This meta-analysis aimed to synthesize available data to verify the association between clinical prognosis value and ROR expression level.
Materials and methods
We performed a systematic search by using PubMed (Medline), Embase, Cochrane Library, ScienceDirect, Springer, and ISI Web of Knowledge from inception to November 15, 2017. Eleven studies with 903 patients were included in this meta-analysis according to the exclusion and inclusion criteria, and the quality of the publications was assessed by using the Newcastle-Ottawa Scale. Pooled odds ratios (OR) and hazard ratios (HR) with 95% CI were used to describe the effect.
Results
The results showed that overexpression of ROR is positively associated with lymph node metastasis (OR=4.472, 95% CI: 3.212–6.225, Z=8.87, P=0.000), tumor invasion depth (OR=9.93, 95% CI: 5.33–18.47, Z=7.24, P<0.001), TNM stage (III/IV vs I/II, OR=2.96, 95% CI: 2.18–4.02, Z=6.95, P<0.001), distant metastasis (OR=3.142, 95% CI: 2.187–4.513, Z=6.20, P<0.001) respectively. Additionally, high expression of ROR was significantly correlated with unfavorable disease-free survival (DFS) (HR=2.74, 95% CI: 1.65–3.82, Z=4.93, P=0.000) and overall survival (OS) (HR=2.09, 95% CI: 1.64–2.54, Z=9.07, P<0.001). Subgroup analysis demonstrated that neither cancer type (digestive or respiratory system) nor sample size (more or less than 100) did not alter the prognostic value of ROR. Furthermore, we performed publication bias and sensitivity analysis in order to examine the stability of meta-analysis of ROR along with OS, which showed that the shape of the funnel plot was nearly symmetrical and the resulting pattern was not significantly influenced while disconnecting each suitable study.
Conclusion
In accordance with these results, we suggested that the overexpression of long noncoding RNA ROR could act as a novel biomarker for predicting poor prognosis in different human cancers.
Introduction
Today, cancer has become one of the major public health concerns globally and the leading cause of death.Citation1 Recent statistics indicate an anticipated 1,688,780 new cases of cancer and approximately 600,920 deaths resulting from cancer in 2017 in the United States. In China, approximately 4,292,000 new cancer cases were estimated to occur along with an estimated 2,814,000 deaths in 2015.Citation2,Citation3 Until now, scientific advancements have allowed us to better understand the molecular mechanism of oncogenesis, progression, and resistance of cancer. However, the evaluation of tumor prognostics, as well as the predictive factors, are still limited to traditional classifications such as anatomic staging, histological typing, and grading. It is widely considered that the new molecular classification algorithm and personalized, accurate medicine are the complements of time-honored classification, primarily as reflected in the eighth edition of AJCC Cancer Staging Manual.Citation4 Therefore, recent studies are devoted to establishing new potential biomarkers for improving the survival of cancer patients.
The human genome is knowingly made up of a vast majority of non-coding RNAs and only approximately 2% protein-coding genes.Citation5 The long non-coding RNAs (lncRNAs) include a group of transcripts in excess of 200 nucleotides with a limited number of protein-coding prospects. They are also deficient in a palpable open reading frame (ORF). Many recent studies have exhibited that lncRNAs might significantly contribute to biological processes such as cellular development and differentiation and to a variety of disease states.Citation6,Citation7 In addition, the deregulation expression of lncRNAs has been shown in multiple types of cancers.Citation8,Citation9 Mechanistic investigation indicated that lncRNAs regulating gene expression through binding to a transcription factor,Citation10 chromatin modifying factorsCitation11,Citation12 or heterogeneous nuclear ribonucleoproteins (hnRNPs)Citation13,Citation14 in the cellular nucleus while acting as an endogenous microRNA “sponge” to take part in the target gene’s post-transcriptional regulation processing in the cytoplasm. In addition, some lncRNAs exhibit distinct developmental, tissue-specific expression patterns and the ability to transduce higher-order spatial information.Citation15–Citation17 All these characteristics made lncRNAs potential for application of cancer patient diagnosis and prognosis, along with serving as a possible therapeutic target.
Long noncoding RNA, regulator of reprogramming (ROR) is 2,591 nts long and is situated at 18q21.31. It also comprises four exons and was first known to promote reprogramming of lineage-committed cells to induced pluripotent stem cells (iPSCs) and maintaining the embryonic stem cells (ESCs) and iPSCs.Citation18 Consequently, Wang et alCitation19 demonstrated that ROR contributes heavily to self-renewal and in the differentiation of human embryonic stem cells (hEScs) as a sponge to miR-145 at the post-transcriptional level. It is also critical in regulating the core transcription factors Oct4, Sox2 and Nanog expression. Further, dysregulation of ROR has been revealed in a variety of cancersCitation20 and primarily implicated in stem-like properties such as EMTCitation21,Citation22 together with chemoresistance.Citation23 Moreover, observations denote that ROR was substantially overexpressed in cancer tissues compared with adjacent non-tumor tissues.Citation24 It was also associated with lymph node metastasis (LNM), histological grade, tumor invasion depth, TNM Stage, overall survival (OS), and disease-free survival (DFS).
However, most articles evaluating the aberrant levels of ROR in cancer are limited in regard to sample sizes and the contentious outcomes. Thus, we performed this meta-analysis to study the overall cancer patient outcome together with the clinical prognostic role of ROR in human cancers.
Materials and methods
Literature search and selection
We carried out the potential eligible literature search in several databases up to November 15, 2017, including PubMed, Embase, Cochrane Library, ScienceDirect, Springer, and ISI Web of Knowledge. The searched keywords were listed as follows: (“LINC-ROR” or “lincRNA-RoR” or “ROR” or “lin-cRNA-ST8SIA3” or “ST8SIA3” or “regulator of reprogramming”) and (“cancer” or “carcinoma” or “tumor” or “tumour” or “neoplasm” or “adenoma” or “sarcoma” or “melanoma”). We also searched the primary literature reference lists and reviews manually to find supplementary pertinent literature. We restricted our literature to English publications.
Inclusion and exclusion criteria
We projected, carried out, and reported this current study in accordance with the PRISMA statement. First, duplicated articles and repetitive data were excluded. The remaining articles were all scanned with titles and abstracts. Excluding tactics were conducted as follows: 1) unrelated to long noncoding RNA ROR, such as retinoic acid receptor-related orphan receptor (ROR), risk of relapse (ROR), radius of rotation (ROR), and reporting odds ratio (ROR); 2) the studies that were only focused on examining the ROR’s structure and functions; 3) non-human investigation, evaluations, letters, case reports, editorials along with specialist opinions; 4) lack of usable clinical data. By contrast, the inclusion criteria were based on: 1) articles examining the clinical functions of long noncoding RNA ROR in different cancers; 2) the patients had been grouped as per the ROR’s expression levels; 3) associated clinic-pathologic factors were defined; 4) description of clinical data, such as LNM, distant metastasis (DM), overall survival (OS), and disease-free survival (DFS); 5) studies with adequate data for the measurement of ORs along with resultant 95% CI or P-value.
Data extraction and quality assessment
Two researchers (SQY and JC) assessed and obtained all the necessary data from ascertained literature autonomously. As per the criteria of inclusion and exclusion, this information was documented: 1) authors, year of publication, and country of origin; 2) cancer type, specimens, and the size of the sample; 3) the technique of ROR assessment; 4) cutoff values and follow-up time; 5) ORs of ROR for the anatomic stage, including LNM, DM, histological grade, tumor invasion depth, TNM stage; and 6) HRs and their corresponding 95% CI of ROR value for OS, DFS. In the case where crucial data were unobtainable from the original literature, efforts were made to get in touch with the consequent author to acquire the raw data. In the case where just the Kaplan–Meier curves had been provided in particular investigations, we extracted the survival rates from the survival plot graphs and the computed HR, 95% CI had been established as the published techniques.Citation25,Citation26 The quality of included publications was evaluated according to the critical checklist of the Dutch Cochrane Center suggested by the Newcastle-Ottawa Scale (NOS). We extracted nine items with each of them scoring 1. The overall scores are in the range of 0–9. A score of ≥7 indicates high-quality study outcome.
Data synthesis and statistical analysis
We collected the pooled HRs and its 95% CI from the included studies depending on which ones were documented in the literature. We used both the log HR along with the standard error (SE) for aggregation of the survival outcome. To assess the heterogeneity of the suitable studies, pooled HRs had been executed by use of I2 statistics in this meta-analysis.Citation27 Then, the random-effects model had been adopted in case of substantial statistical heterogeneity among the studies (chi-squared test, P<0.1) in analyzing the results. Where not applicable, we used the fixed-effects model (chi-squared test, P>0.1). We applied forest plots to display the meta-analysis outcomes. We further assessed any prospective bias in the publication by use of the Egger’s test. We conducted all the statistical analyses by use of the stata12.0 (StataCorp, USA). Additionally, we regarded P<0.05 to be statistically significant.
Results
Characteristics of the included studies
The flow diagram in illustrates a total of 760 published records that were carried out in the preliminary search and another 273 duplicate articles that were disregarded. Subsequent to careful inspection of the title and abstract, a total of 426 inappropriate articles were omitted while 61 prospective and suitable studies were selected. Following a further review of these articles, 15 had been excluded for lacking clinical information, whereas 10 others had been considered not dissenting into the high and low ROR expression groups. Additionally, 16 of the articles did not have data on the OS or DFS outcomes, while nine were found to lack sufficient information needed for calculation of OR and 95% CI or P-value. Ultimately, as per the selection criteria, a total of 11 articles were found suitable for further research.Citation28–Citation38.
These studies were mainly published from 2016–2017. Ten of the studies included were obtained from China, whereas one was from India. Out of the 11 studies, two emphasized non-small-cell lung cancer (NSCLC), while two others were based on pancreatic cancer (PC). Additionally, one of them focused on gastric cancer (GC), with another one focusing on esophageal squamous cell carcinoma (ESCC). There was also one based on hepatocellular carcinoma (HCC), another one taking into account colon cancer (CC), one on gallbladder cancer (GAC), another on bladder cancer (BLC), and the last one emphasized oral cancer. We detected the expression of ROR and normalized qRT-PCR to GAPDH or β-actin. For all of the investigations, we put the patients into two distinct groups: high and low expression of ROR. We also extracted HR and its 95% CI directly from the original information reported in the literature. We based all the diagnoses on pathology. summarizes the main features of the suitable studies. The NOS affirmed that all of these studies were of the required high quality as shown in .
Table 1 Characteristics of the included studies in this meta-analysis
Table 2 Quality assessment of eligible studies (Newcastle-Ottawa Scale)
Meta-analysis
ROR and tumor histological grade
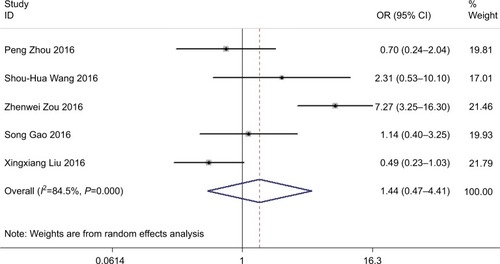
We included a total of five studies that reported an overall tumor histological grade on the basis of varied ROR expression levels. We further used the random-effects model as the significant heterogeneity (I2=84.5%, P<0.001). Analysis exhibited that pooled OR was 1.445 with 95% CI: 0.473–4.409 (Z=0.65, P=0.518) (). The result indicated that 95% CI crossed 1, meaning that there was no significant differentiation. Owing to heterogeneity, we carried out a sensitivity analysis to ascertain the potential source of heterogeneity. After disregarding the Zou,Citation31 the heterogeneity was observed to apparently decline (I2=28%, P=0.25) and OR=0.82 with 95% CI: 0.45–1.49 (Z=0.66, P=0.51) (data not shown). There was also no significant difference in the tumor histological grade between these two groups.
ROR and tumor LNM
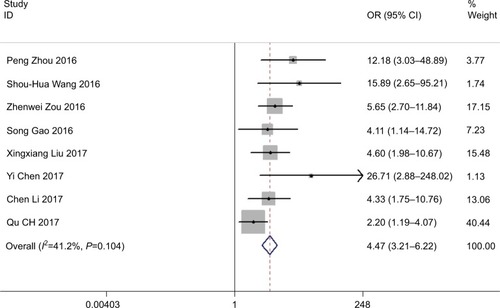
We included an aggregate of patients in eight studies to distinguish the connection between ROR expression levels and the tumor lymph node metastasis. We observed a substantial relationship between LNM and ROR expression (OR=4.472, 95% CI: 3.212–6.225, Z=8.87, P=0.000) (). Then, we applied the fixed-effects Mantel–Haenszel model for the heterogeneity test (I2=41.2%, P=0.104). The outcomes confirmed a significant difference in the LNM incidence between the two groups while demonstrating that high ROR expression may significantly perceive a higher tendency of developing LNM in cancer patients.
ROR and tumor invasion depth
Four studies reported the depth of tumor invasion on the basis of the various ROR levels of expression. The investigation demonstrated a significant association between tumor invasion depth and ROR expression with a pooled OR=5.77 (95% CI: 1.58–21.07, Z=2.65, P=0.008) in a random-effects model (). Due to the presence of heterogeneity (I2=83.2%, P<0.001), we conducted a sensitivity analysis to simplify the probable source of heterogeneity. We found that the heterogeneity visibly dropped (I2=39.4%, P=0.176) without a change in the result (OR=9.93, 95% CI: 5.33–18.47, Z=7.24, P<0.001) following exclusion of the Liu studyCitation36 (). The result also confirmed a significant difference in the tumor invasion depth in the two groupings. Thus, the results showed that high ROR expression might significantly perceive a higher depth of tumor invasion in cancer patients.
ROR and tumor TNM stage
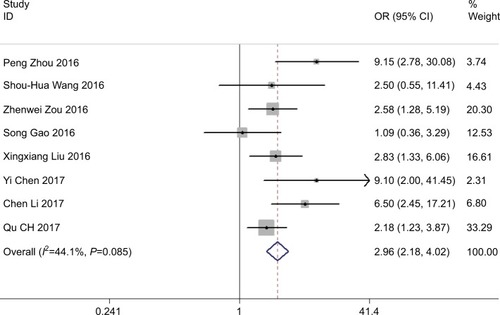
We further included the patients in the eight studies to distinguish the association between high and low expression of ROR. We performed a fixed-effects model to compute the pooled OR 2.96 and its 95% CI (2.18–4.02) (Z=6.95, P<0.001) when no observable heterogeneity in studies (I2=44.1%, P=0.085). indicates that the elevated ROR expression significantly detected poor TNM stage III/IV.
ROR and tumor DM
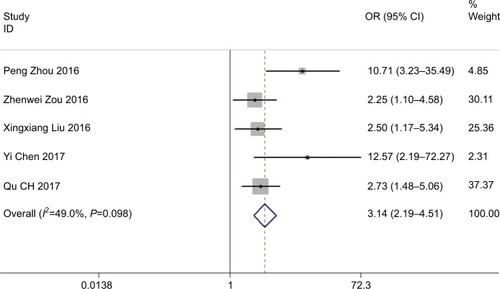
A total of five studies reported the tumor DM on the basis of varied levels of ROR expression. The analysis revealed a pooled OR=3.142 (95% CI: 2.187–4.513, Z=6.20, P<0.001) in a fixed-effects model (I2=49.0%, P=0.098) (). The outcome showed that overexpression of ROR was significantly associated with DM.
Association between ROR and DFS
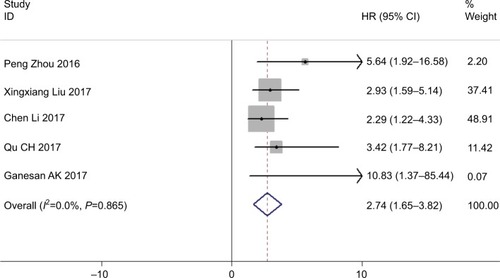
We included patients in five studies to investigate the association between levels of ROR expression and patients’ DFS. We detected a significant relationship between DFS and ROR expression (HR =2.74, 95% CI: 1.65–3.82, Z=4.93, P=0.000) (). Additionally, we did not identify any significant heterogeneity among the studies (I2=0.0%, P=0.865), demonstrating that high ROR expression might be significantly related to shorter DFS.
Association between ROR and OS including subgroup analysis
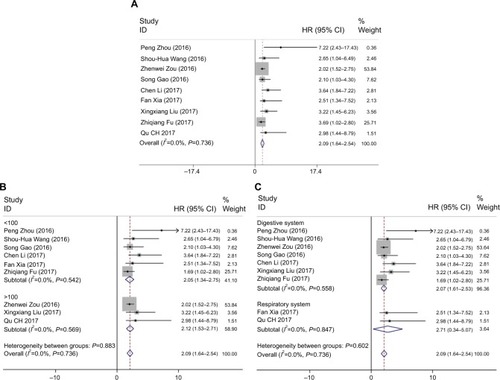
We also collected pooled HRs data and 95% CI of OS from the nine suitable studies. The analysis revealed a pooled HR of 2.09 with 95% CI (1.64–2.54) (Z=9.07, P<0.001), through a fixed-effects model (I2=0.0%, P=0.736) (). Further, we performed a subgroup analysis to examine the sources of heterogeneity. In a subgroup analysis of the sample size, there was an observable significant association between high ROR expression levels and OS both fewer than 100 (HR: 2.05, 95% CI: 1.34–2.75, Z=5.70, P<0.001) and more than 100 (HR: 2.12, 95% CI: 1.53–2.71, Z=7.06, P<0.001). It was also observable that the relationship between ROR and OS was significant in studies with digestive system cancers (HR: 2.07, 95% CI: 1.61–2.53, Z=8.81, P<0.001) and patients with respiratory system cancer (HR: 2.71, 95% CI: 0.34–5.071, Z=2.24, P=0.025). However, there was no significant heterogeneity in this subgroup analysis ().
Publication bias and sensitivity analysis
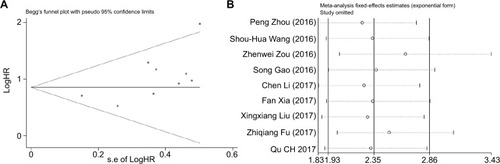
We evaluated the publication bias of this meta-analysis through the funnel plot and the Egger’s test. It was observable that the shape of the funnel plot was nearly symmetrical, which did not show any indication of apparent asymmetry (), Begg’s test P=0.118. The publication bias for LNM, TNM, DM and DFS groupings was not studied due to the small number of studies or possible lack of heterogeneity. We further undertook a sensitivity analysis in order to examine the stability of the meta-analysis of ROR along with OS by successively disconnecting each suitable study. Also, the resulting pattern was not significantly influenced (). In accordance with these results, we realized that overexpression of ROR could predict poor prognosis in different cancers ().
Table 3 A summary of the results of this meta-analysis
Discussion
The classical central dogma cannot sufficiently elucidate the underlying genetic complexity associated with higher eukaryote after many crucial, highly conserved metabolic and environmental protein-based feedback loops have been studied. This complexity is most likely derived from the noncoding RNA (ncRNA), once thought of as “junk DNA.”Citation39 Research over the last decade revealed a new class of transcripts, known as long noncoding RNAs (lncRNAs). Moreover, various lines of proof have continually linked mutations and dysregulations of lncRNAs to different human diseasesCitation40 such as cancer.Citation9 Many studies demonstrated that lncRNAs show spatial-, temporal-, and tissue-specific and cell-state patterns of expressionCitation16,Citation17,Citation41 and thus might operate as an oncogene or tumor suppressor through regulating survival, proliferation, invasion, metastasis and angiogenesis of cancer cells.Citation8 Additionally, the lncRNAs have been identified within cells and in biological fluids such as serum, plasma, urine, and saliva of cancer patients.Citation8 Collectively suggesting that lncRNA can be used as a diagnostic and prognostic biomarker.Citation42–Citation44
Long noncoding RNA ROR, one of the reprogramming-stimulated lncRNAs, is precisely targeted by main pluripotency aspects and modulates reprogramming or iPSC maintenance.Citation18 Knockdown of ROR substantially decreased the efficiency of reprogramming, while its overexpression brought about a higher than two-fold upsurge in the formation of iPSC colony. Microarray gene expression profiling proposed that the knockdown of ROR caused an upregulation of genes implicated in cellular stress pathways and p53 response, whereas p53 plays an important role in stem cell and cancer biology. Subsequently, underlying mechanism studies found that ROR contributes to the oncogenic role partly through suppressing p53 translation and promoting the stability of c-Myc mRNA through direct interaction with the heterogeneous nuclear ribonucleoprotein (hnRNP) I.Citation45,Citation46 In addition, studies demonstrated that ROR also maintain stem cell pluripotency via sponging miRNAs and promote tumorigenesis, thereby inducing the epithelial–mesenchymal transition (EMT) in the cancer cells.Citation21,Citation22,Citation47–Citation49 Additionally, numerous studies have verified ROR aberrant overexpression in several types of cancers tissue.Citation21–Citation23,Citation28–Citation38,Citation46,Citation50 However, Feng et alCitation51 reported that the expression of ROR was decreased in 23 glioma specimens and elevated in the other three specimens compared to matched noncancerous tissue with no complete clinical data. Subsequently, gain and loss of function studies have shown that overexpressed ROR could inhibit glioma cell line U87 proliferation, suggesting that ROR may serve as a novel tumor suppressor gene in glioma. Previous studies have revealed that ROR maintains cancer stem cell self-renewal and undifferentiated state,Citation18,Citation45,Citation46,Citation52 this indicated that ROR expression might be restricted to stem-like properties and a less differentiated population of cancer cells, which may highlight the explanation of the downregulation result occurring in cell lines and tissue specimens. Moreover, we generally observed the detection of gene expression was varied between the individual samples, calling for a large number cohort of cancer samples to be analyzed. Nevertheless, all collections indicated that long noncoding RNA ROR might exert useful diagnostic and prognostic biomarker.
This meta-analysis aimed at exploring the evidence of whether high expression of ROR contributes to an induced poor prognosis in patients with cancer. We consider that this meta-analysis is the first to provide comprehensive and detailed discernments into the clinical significance of long noncoding RNA ROR in carcinomas. In this study, we pooled eleven studies, including 903 patients, in which the result proposed that elevated ROR expression is observably correlated to poor prognosis, progression, primarily in LNM, tumor invasion depth, TNM stage, and DM, together with DFS and OS. Our analysis illustrated that a pooled HR was 4.47 (95% CI: 3.21–6.22, P<0.001), 9.93 (95% CI: 5.33–18.47, P<0.001), 2.96 (95% CI: 2.18–4.02, P<0.001), and 3.14 (95% CI: 2.19–4.51, P<0.001) for LNM, tumor invasion depth, TNM stage, and DM correspondingly. However, there is no significant difference in tumor histological grade and the pooled HR 0.77 with 95% CI: 0.47–1.26 crossed (P=0.294).
We observed significant heterogeneity between ROR expression and tumor invasion depth (I2=83.2%, P<0.001). Thus, heterogeneity can be identified to be a likely problem in interpreting the outcomes of any meta-analysis. To resolve the issue, we carried out a sensitivity analysis in which the existent heterogeneity palpably reduced (I2=39.4%, P=0.176) after exclusion of the Liu study.Citation36 However, the results did not vary (OR=9.93, 95% CI: 5.33–18.47, Z=7.24, P<0.001). It means that Liu’s study might have a substantial impact on the overall heterogeneity. In our broadened analysis, we assumed the following: First, the prospective sources of heterogeneity for the result to encompass the sample size and a sample selection of the study might alter the heterogeneity. Second, the qRT-PCR reaction conditions, along with the reaction systems, were varied in the involved studies, thus influencing the outcome. Additionally, the features of various tumor clinical pathological characteristics may cause possible heterogeneity. This paper considers that the pathological type of esophageal squamous cell carcinoma might cause heterogeneity. Lastly, the variable cutoff value for ROR in this study may be associated with the heterogeneity.
Additionally, through merging HRs from the Cox multivariate studies, we detected that ROR was an independent prognostic factor of DFS and OS for patients with cancer (pooled HR 2.74, 95% CI: 1.65–3.82 and 2.09, 95% CI: 1.64–2.54 respectively). In the stratified analysis, the outcomes proposed that elevated levels of ROR could significantly associate with a worse result regardless of being in the digestive system (HR: 2.09, 95% CI: 1.64–2.54, P<0.001) or non-digestive system (HR: 2.71, 95% CI: 0.34–5.071, P=0.025). The relationship between ROR and OS was found to be substantial in studies with fewer sample sizes of 100 (HR: 2.05, 95% CI: 1.34–2.75, P<0.001) and greater than 100 (HR: 2.12, 95% CI: 1.53–2.71, P<0.001). Additionally, we detected a substantial relationship between MFS and ROR expression (HR: 2.74, 95% CI: 1.65–3.82, P=0.000).
Limitations
However, this study has a number of limitations: 1) all the articles that were included were conservative, whereas some of them had small sample sizes; only eleven studies with 9 types of tumors had been incorporated into the meta-analysis; 2) specific HR had been computed through reconstruction of survival curves instead of precisely obtaining them from the primary studies; 3) the cutoff values vary among studies that might have weakened the reliability of the conclusion; 4) the primary patients who were included in the study were from China, with only one from India. Thus, the findings might just be representative of Asian patients. Notably, different types of cancer specimen sources may show inherent molecular differences and increased the heterogeneity. Thus, it is strongly recommended that a larger-size and better design study is carried out to substantiate these results.
Conclusion
Notwithstanding the limitations stated earlier, this meta-analysis provides a preliminary indication that higher ROR expression might be regarded as a reliable uncomplementary prognostic factor in human cancers. High ROR expression is associated with increased tumor invasion depth, LNM, TNM stage, poor DFS and OS. In future clinical studies, it is relevant to use a better design in verifying and strengthening the current prognostic biomarker role of long noncoding RNA ROR in patients with neoplasm.
Acknowledgments
The present study was financially supported by funds from the National Natural Science Foundation of the People’s Republic of China (8177235, 81520108018 and 81472080), the Jiangsu Committee of Science and Technology-Social Development Plan (BE2017755), and the Nanjing Committee of Science and Technology (201505005).
Author contributions
SQY, JC and YY contributed to the conception of the study, participating in its design, analyzing the data, and coordinating to draft the manuscript, these authors have contributed equally to this work. DLL, MYH participated in the design of the study, searched and selected the trials and performed the statistical analysis. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- TorreLABrayFSiegelRLFerlayJLortet-TieulentJJemalAGlobal cancer statistics, 2012CA Cancer J Clin20156528710825651787
- ChenWZhengRBaadePDCancer statistics in China, 2015CA Cancer J Clin201666211513226808342
- SiegelRLMillerKDJemalACancer statistics, 2017CA Cancer J Clin201767173028055103
- AminMBGreeneFLEdgeSBThe eighth edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer stagingCA Cancer J Clin2017672939928094848
- UlitskyIBartelDPlincRNAs: genomics, evolution, and mechanismsCell20131541264623827673
- FaticaABozzoniILong non-coding RNAs: new players in cell differentiation and developmentNat Rev Genet201415172124296535
- SunMKrausWLFrom discovery to function: the expanding roles of long noncoding RNAs in physiology and diseaseEndocr Rev2015361256425426780
- HuarteMThe emerging role of lncRNAs in cancerNat Med201521111253126126540387
- EvansJRFengFYChinnaiyanAMThe bright side of dark matter: lncRNAs in cancerJ Clin Invest201612682775278227479746
- HuXFengYZhangDA functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancerCancer Cell201426334435725203321
- GuptaRAShahNWangKCLong non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasisNature201046472911071107620393566
- ZhangEHanLYinDH3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinomaNucleic Acids Res20174563086310127956498
- LiZChaoTCChangKYThe long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPLProc Natl Acad Sci U S A201411131002100724371310
- AtianandMKHuWSatpathyATA long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammationCell201616571672168527315481
- MercerTRDingerMESunkinSMMehlerMFMattickJSSpecific expression of long noncoding RNAs in the mouse brainProc Natl Acad Sci U S A2008105271672118184812
- GrotePWittlerLHendrixDThe tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouseDev Cell201324220621423369715
- FlynnRAChangHYLong noncoding RNAs in cell-fate programming and reprogrammingCell Stem Cell201414675276124905165
- LoewerSCabiliMNGuttmanMLarge intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cellsNat Genet201042121113111721057500
- WangYXuZJiangJEndogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewalDev Cell2013251698023541921
- QuekXCThomsonDWMaagJLlncRNAdb v2.0: expanding the reference database for functional long noncoding RNAsNucleic Acids Res201543Database issueD168D17325332394
- HouPZhaoYLiZLincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasisCell Death Dis20145e128724922071
- ZhanHXWangYLiCLincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathwayCancer Lett2016374226127126898939
- LiLGuMYouBLong non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinomaCancer Sci201610791215122227311700
- RezaeiMEmadi-BaygiMHoffmannMJSchulzWANikpourPAltered expression of LINC-ROR in cancer cell lines and tissuesTumour Biol20163721763176926314857
- ParmarMKTorriVStewartLExtracting summary statistics to perform meta-analyses of the published literature for survival endpointsStat Med19981724281528349921604
- TierneyJFStewartLAGhersiDBurdettSSydesMRPractical methods for incorporating summary time-to-event data into meta-analysisTrials200781617555582
- IoannidisJPPatsopoulosNAEvangelouEHeterogeneity in meta-analyses of genome-wide association investigationsPLoS One200729e84117786212
- GaoSWangPHuaYROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancerOncotarget2016721608161826636540
- WangSHZhangMDWuXCWengMZZhouDQuanZWOverexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasionTumour Biol2016379128671287527449039
- ZhouPSunLLiuDLiuCSunLLong non-coding RNA lincRNA-ROR promotes the progression of colon cancer and holds prognostic value by associating with miR-145Pathol Oncol Res201622473374027071407
- ZouZDingQLiPOverexpression of lincRNA-ROR predicts poor prognosis in patients with gastric cancerInt J Clin Exp Pathol20169994679472
- ArunkumarGDeva Magendhra RaoAKManikandanMExpression profiling of long non-coding RNA identifies linc-RoR as a prognostic biomarker in oral cancerTumour Biol2017394101042831769836628443494
- ChenYPengYXuZLncROR promotes bladder cancer cell proliferation, migration, and epithelial-mesenchymal transitionCell Physiol Biochem20174162399241028463831
- FuZLiGLiZEndogenous miRNA sponge LincRNA-ROR promotes proliferation, invasion and stem cell-like phenotype of pancreatic cancer cellsCell Death Discov201731700428580169
- LiCLuLFengBThe lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2Sci Rep201771463728680141
- LiuXCuiLLiuJIncreased LincRNA ROR is association with poor prognosis for esophageal squamous cell carcinoma patientsInt J Clin Exp Pathol201710446544660
- QuCHSunQYZhangFMJiaYMLong non-coding RNA ROR is a novel prognosis factor associated with non-small-cell lung cancer progressionEur Rev Med Pharmacol Sci201721184087409129028092
- XiaFXiongYLiQInteraction of lincRNA ROR and p53/miR-145 correlates with lung cancer stem cell signaturesJ Cell Biochem Epub ahead of print
- AdamsBDParsonsCWalkerLZhangWCSlackFJTargeting noncoding RNAs in diseaseJ Clin Invest2017127376177128248199
- WapinskiOChangHYLong noncoding RNAs and human diseaseTrends Cell Biol201121635436121550244
- SauvageauMGoffLALodatoSMultiple knockout mouse models reveal lincRNAs are required for life and brain developmentElife20132e0174924381249
- GutschnerTDiederichsSThe hallmarks of cancer: a long non-coding RNA point of viewRNA Biol20129670371922664915
- QiPduXThe long non-coding RNAs, a new cancer diagnostic and therapeutic gold mineMod Pathol201326215516522996375
- Chandra GuptaSNandan TripathiYNandanTYPotential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targetsInt J Cancer201714091955196727925173
- ZhangAZhouNHuangJThe human long non-coding RNA-RoR is a p53 repressor in response to DNA damageCell Res201323334035023208419
- HuangJZhangAHoTTLinc-RoR promotes c-Myc expression through hnRNP I and AUF1Nucleic Acids Res20164473059306926656491
- ZhangYXiaJLiQNRF2/long noncoding RNA ROR signaling regulates mammary stem cell expansion and protects against estrogen genotoxicityJ Biol Chem201428945313103131825231996
- ZhouXGaoQWangJZhangXLiuKDuanZLinc-RNA-RoR acts as a “sponge” against mediation of the differentiation of endometrial cancer stem cells by microRNA-145Gynecol Oncol2014133233333924589415
- PanYChenJTaoLLong noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathwayOncotarget2017820331443315828388536
- PengWXHuangJGYangLGongAHMoYYYyMLinc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancerMol Cancer201716116129041978
- FengSYaoJChenYExpression and functional role of reprogramming-related long noncoding RNA (lincRNA-ROR) in gliomaJ Mol Neurosci201556362363025651893
- FanJXingYWenXLong non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesisGenome Biol20151613926169368