Abstract
Purpose
This study aimed to study the biological function and the molecular mechanisms associated with the promotion of angiogenesis by lncRNA LOC100132354 in lung adenocarcinoma (LAD).
Patients and methods
The mRNA expression levels of 100 pairs of LAD and normal tissue samples of LOC100132354, vascular endothelial growth factor A (VEGFA), VEGF receptor-2 (VEGFR2), basic fibroblast growth factor (bFGF), and thrombospondin-1 (TSP-1) were analyzed by qPCR. LOC100132354 was knockdown and overexpressed in SPCA-1 and A549 cell lines to analyze the protein and mRNA expression levels of VEGFA, VEGFR2, bFGF, TSP-1, and changes in protein expression levels of Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2. Tumor microvessel density (MVD) was analyzed in experimental nude mice.
Results
The qPCR results showed that the mRNA expression levels of LOC100132354, VEGFA, VEGFR2, and bFGF mRNA in LAD tissues were significantly increased, while TSP-1 mRNA was significantly decreased compared with the adjacent tissues. Survival analysis showed that VEGFA, VEGFR2, and bFGF were poor predictors, while TSP-1 was a good predictor in LAD. Knockdown or overexpression of LOC100132354 affected the expression levels of bFGF, VEGFA/VEGFR2 signaling pathway, and downstream target molecules, such as Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2, while decreased TSP-1. After knockdown or overexpression of VEGFA expression, no significant changes in the expression level of LOC100132354 were found. Tumorigenesis of nude mice confirmed that LOC100132354 can significantly increase the tumor MVD.
Conclusion
These findings suggest VEGFA was a downstream target gene of LOC100132354, promoting angiogenesis through VEGFA/VEGFR2 signaling pathway and downstream target molecules in LAD. So, LOC100132354 is considered as an antiangiogenic target in LAD.
Introduction
Lung cancer ranks first in terms of both morbidity and mortality of all cancers.Citation1 Of all lung cancers, non-small-cell lung cancer (NSCLC) accounts for approximately 85%. According to the histopathological classification, NSCLC is divided into lung adenocarcinoma (LAD) and squamous cell carcinoma. Despite several advancements in chemotherapy, radiation, and surgery, lung cancer still remains aggressive and resulted in mortality.Citation2 The average 5-year survival of lung cancer is below 15%.Citation3–Citation6 Recently, an increasing proportion of LAD has drawn a great attention on the socioeconomic environments.Citation7 However, the mechanisms of LAD have not yet been uncovered.
LncRNAs have been confirmed to play a very important role in the occurrence and progression of lung cancer. It has been found that some lncRNAs, such as HOTAIR,Citation8 H19,Citation9 MALAT1,Citation10 SCAL1,Citation11 AK126698,Citation12 and GAS6-AS1,Citation13 are closely related to lung cancer. Previously, we have found that lncRNA LOC100132354 was overexpressed in LAD and was localized at 6p21.1, with a total of 2282 bp RNA sequences encompassing the two exons by gene microarray. With these results, we have confirmed that LOC100132354 is an oncogenic lncRNA gene, and it promoted the progression and markedly enhanced the oncogenic ability in LAD.Citation14 Therefore, we performed bioinformatics analysis of LOC100132354 and its adjacent coding gene (distance <300 kb) with transcriptome mRNA microarray results. These findings revealed that vascular endothelial growth factor A (VEGFA) may be an adjacent gene regulated by LOC100132354.
The growth and metastasis of lung cancer require the support of new blood vessels. Neovascularization plays a key role in the occurrence and development of lung cancer and is determined by the tumor microenvironment. Multiple signaling molecules were involved in its regulation.Citation15–Citation19 The proliferation of vascular endothelial cells is considered to be the core of neovascularization process. Vascular endothelial growth factor (VEGF) is the most important factor for inducing endothelial cell proliferation, and angiogenesis in tumor tissues is directly related to VEGF. VEGF consists of five family members, namely VEGFA, VEGFB, VEGFC, VEGFD, and VEGFE. VEGF binds to its receptors to induce the biological effects. The VEGF receptors (VEGFRs) mainly include Flt-1, KDR/Flk-1 (VEGFR2), Flt-4, and so on.Citation20,Citation21 VEGF, VEGFR, and their downstream signaling transduction pathways are used as specific tumor angiogenesis markers. Also VEGFA/VEGFR2 signaling transduction plays a leading role in the formation of vascular endothelial cells. VEGFA/VEGFR2 is a key factor for promoting angiogenesis in lung cancer,Citation22 and it can promote tumor growth.Citation23 Therefore, inhibition of VEGFA/VEGFR2 signaling pathway can inhibit tumor cell growth by blocking angiogenesis.Citation24,Citation25 Basic fibroblast growth factor (bFGF) is also one of the angiogenic growth factors that can stimulate endothelial cells to synthesize collagenase and promote invasion of the surrounding stroma during angiogenesis. Moreover, VEGF can induce endothelial cell production and release bFGF, further promoting the role of angiogenesis.Citation26 While throm-bospondin-1 (TSP-1) is a strong negative regulator of tumor angiogenesis.Citation27
VEGFA/VEGFR2 can activate Ras by multiple pathwaysCitation28 and in turn activate Raf/Mek/Erk to exert the biological functions. Studies have shown that specific inhibitors of Mek1/2 can prevent the activation of Erk, thereby inhibiting the formation of blood vessels in vitro. Therefore, we speculate that LOC100132354 may regulate VEGFA/VEGFR2 signaling pathway to promote angiogenesis in LAD, but the specific regulatory mechanisms needs to be elucidated.
Patients and methods
Patient samples
We collected 100 LAD tissue samples and corresponding normal tissue (NT) samples in the First Affiliated Hospital of Wenzhou Medical University, from January 2013 to August 2015. The TNM stage was determined according to the American Joint Committee on Cancer (AJCC) and the Union for International Cancer Control (UICC) in 2002. The diagnosis of LAD was confirmed using histopathology results. The clinical and characteristic data of these LAD patients are presented in . We followed-up the survival of these cases by telephone or other ways, and the range of follow-up time was 3–23 months. This study was approved by the Institutional Ethics Review Board of the First Affiliated Hospital of Wenzhou Medical University, and all patients provided written informed consent.
Table 1 The clinical features of 100 patients with LAD
Quantitative PCR
Total RNA was obtained from fresh LAD tissues and cell lines with TRIzol Reagent kit (Invitrogen, Waltham, MA, USA) and was reverse-transcribed into cDNA with a reverse-transcribed kit (TaKaRa, Shanghai, China) according to the manufacturer’s instructions. LOC100132354, VEGFA, VEGFR2, bFGF, TSP-1, and β-actin mRNA expressions were detected by qPCR in ABI 7300 analyzer. The primers of six genes are shown in . Two micrograms of total RNA was reverse-transcribed into cDNA. qPCR was performed in a total reaction volume of 20 µL, which consists of 2 µL cDNA, 1 µL PCR forward primer (10 mM), 10 µL SYBR Premix (2×), 1 µL PCR reverse primer (10 mM), and 6 µL distilled water. The qPCR program was as follows: 10 minutes denaturation at 95°C, 40 cycles for 5 seconds at 95°C, 30 seconds at 60°C, and 5 minutes extension at 72°C. All the results were normalized to β-actin, and all the experiments were performed in triplicate. The median in each triplicate for calculating the relative lncRNA expressing concentrations (DCt=Ct median Ct median β-actin), and 2−ΔΔCt formula for the relative expression.Citation29
Table 2 The primers of five genes
Cell culture
Two human LAD cell lines (SPCA-1, A549) were purchased from the Cell Bank of the Chinese Academy of Sciences and were cultured with complete medium (90% RPMI1640 supplemented with 10% FBS) at 37°C in 5% CO2. The complete medium was replaced every 2–3 days.
Lentivirus-mediated siRNA and overexpression vector construction
We constructed siRNA GV248 vector and overexpression GV303 vector for LOC100132354 (GeneChem, Shanghai, China). SiRNA sequences of LOC100132354 were as follows: siRNA1: CCACCTGGATAGGGACAAA, siRNA2: GAAGAGAAATTGACAGGTT, siRNA3: CTGG AGTAGAGAAGA GAAT. SiRNA sequences of VEGFA were as follows: siRNA1: AAGCCGTCCT GTGTGC-CGCTG, siRNA2: AACGATGAAGCCCTGGAGTGC, siRNA3: AAG TTAAAAGTGCCTGAACTG. Transfection was performed by adding 2×105 cells into 6-well plate. After 24 hours, the medium was changed, and the cells were incubated with the transfection complex according to the manufacturer’s standard operating procedure. The multiplicity of infection valuesCitation30 was as follows: the MOI of A549 was 20 and that of SPCA-1 was 100. The A549 and SPCA-1 cells were infected with these lentiviruses for 72 hours, and the siRNA or overexpression efficiency was evaluated by qPCR. Puromycin screen experiment separated and obtained these cell lines by successfully transfecting with lentivirus-mediated vectors. The study involved SPCA-1 LOC100132354, VEGFA siRNAs cell lines (siRNAs group, including siRNA1, siRNA2, and siRNA3). SPCA-1 was transfected with lentivirus-negative control LVCON077 vector (siRNA NC group), SPCA-1 (siRNA control group), and the A549 LOC100132354. VEGFA overexpressing cell line (OE group) and A549 were transfected with lentivirus negative control LVCON145 vector (OE NC group) and A549 (OE control group). We have previously constructed the best LOC100132354 interference sequence for SPCA-1 cell line and LOC100132354 overexpression level in A549 cells.Citation14
Western blotting
The cell lysate was placed on ice for 30 minutes. The supernatant was electrophoresed on SDS-PAGE and then transferred onto a nitrocellulose membrane. Following the molecular cloning procedure, the primary antibody was blocked overnight and horseradish peroxidase secondary antibody was allowed to bind for 1 hour.Citation31 The protein levels of VEGFA, VEGFR2, bFGF, TSP-1, Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2 in these groups were then analyzed.
Nude mice tumor formation and microvessel density (MVD) detection
The cells include SPCA-1 LOC100132354 siRNA2 cell line (siRNA group), siRNA NC group, OE group, and OE NC group. These cells at a density of 2×106 cells were inoculated into the axillary region of BALB/c nude mice subcutaneously. MVD was measured according to the method reported by Weidner et al and Maeda et al, and the sections were analyzed to determine the highest blood vessel density in the tumor, necrosis area with lesser blood vessels, and no blood vessels at the junction with NTs under low magnification microscope (100×). Any brown-yellow stained endothelial cells or cell plexuses that distinguish from the surrounding tumor tissues and connective tissue components were considered as blood vessels and confirmed after finding the highest density of blood vessels under the high magnification microscope (200×). If the structures are not connected, its branching structure also serves as a blood vessel.Citation32,Citation33 The number of microvessels of the five fields of vision was recorded for each sample, and the average number of groups was taken as the MVD value for the group. The investigation confirms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85–23, revised 1996). The study protocol was approved by the Institutional Animal Experimental Ethics Review Board of Wenzhou Medical University.
Statistical methods
Differences in variables between the groups were assessed using Kruskal–Wallis test for non-normal distribution data or one-way ANOVA test for normal distribution data. Comparison between the two groups was evaluated by Student’s t-test or Mann–Whitney U test or least significant difference test. Survival curve was analyzed with GraphPad Prism software 6.0, and chi-squared test analyzed the differences. P<0.05 was considered to be statistically significant.
Results
The expression level and survival analysis of LOC100132354, VEGFA, VEGFR2, bFGF, and TSP-1 in LAD
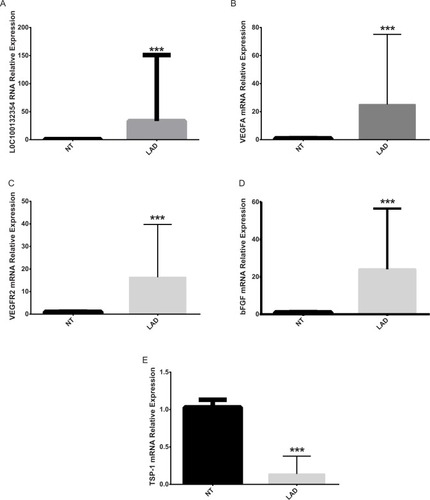
The results of qPCR showed that the relative mRNA expression level of LOC100132354, VEGFA, VEGFR2, and bFGF was obviously higher than the NT tissues (Mann–Whitney U=103.00, 87.00, 79.00, 96.00 and P=0.0040, 0.0018, 0.0019, 0.0021), while TSP-1 mRNA was obviously lower than the NT tissues (Mann–Whitney U=165.00, P=0.0000) in LAD (). This suggested significantly enhanced angiogenic capacity in the LAD tissues. One-way ANOVA test or t-test showed that the relative mRNA expression levels of VEGFA, VEGFR2, bFGF, and TSP-1 (P=0.001, 0.008, 0.019, 0.021. 0.008, 0.002, 0.021, 0.032. 0.000, 0.000, 0.002, 0.012) were obviously different in clinical stage, differentiation degree, and lymph node metastasis, while VEGFA, VEGFR2, bFGF, and TSP-1 (P=0.876, 0.765, 0.543, 0.762. 0.854, 0.734, 0.521, 0.773) showed no differences in sex and smoking ().
Figure 1 Comparison of mRNA expression levels of LOC100132354, VEGFA, VEGFR2, bFGF, and TSP-1 in lung adenocarcinoma tissues and adjacent tumors.
Notes: (A) Comparison of the relative expression levels of LOC100132354 in LAD and adjacent tissues showed that LOC100132354 expression level in LAD was significantly higher than the adjacent tissues. ***P<0.001. (B) Comparison of the relative expression levels of VEGFA in LAD and adjacent tissues showed that VEGFA expression level in LAD was distinctly higher than the adjacent tissues. ***P<0.001. (C) Comparison of the relative expression levels of VEGFR2 in LAD and adjacent tissues showed that VEGFR2 expression level in LAD was evidently higher than the adjacent tissues. ***P<0.001. (D) Comparison of the relative expression levels of bFGF in LAD and adjacent tissues showed that bFGF expression level in LAD was obviously higher than the adjacent tissues. ***P<0.001. (E) Comparison of the relative expression levels of TSP-1 in LAD and adjacent tissues showed that TSP-1 expression level in LAD was markedly lower than the adjacent tissues. ***P<0.001.
Abbreviations: bFGF, basic fibroblast growth factor; LAD, lung adenocarcinoma; NT, normal tissue; TSP-1, thrombospondin-1; VEGFA, vascular endothelial growth factor A; VEGFR2, vascular endothelial growth factor receptor-2.
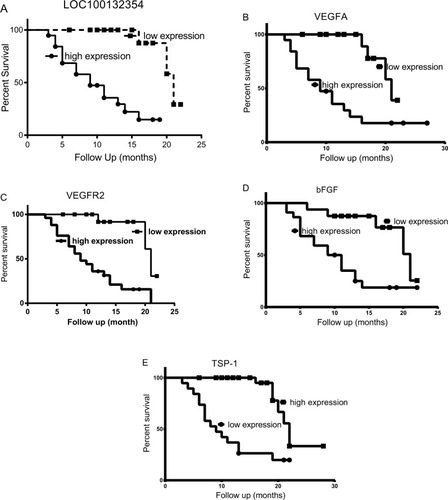
The survival analysis showed that VEGFA, VEGFR2, and bFGF (χ2=16.26, 10.78, 16.22 and P<0.0001, P=0.0010, P<0.0001) were considered as poor predictors, while TSP-1 (χ2=17.50, P<0.0001) was a good predictor in LAD (). Studies have confirmed that VEGFA and VEGFR2 played a pivotal role in angiogenesis and bFGF was considered as an angiogenic factor, while TSP-1 was a negative regulator of vascular growth. So, our results confirmed that the growth of blood vessels was significantly increased in the LAD tissues.
Figure 2 The results of survival analysis in LAD.
Notes: (A) The survival analysis showing that LOC100132354 (χ2=12.22, P<0.001) as a poor predictor. (B) The survival analysis showing VEGFA (χ2=16.26, P<0.0001) as a poor predictor. (C) The survival analysis showing that VEGFR2 (χ2=10.78, P=0.0010) as a poor predictor. (D) The survival analysis showing that bFGF (χ2=16.22, P<0.0001) as a poor predictor. (E) Survival analysis showing TSP-1 as a good predictor in LAD (χ2=17.50, P<0.0001).
Abbreviations: bFGF, basic fibroblast growth factor; LAD, lung adenocarcinoma; TSP-1, thrombospondin-1; VEGFA, vascular endothelial growth factor A; VEGFR2, vascular endothelial growth factor receptor-2.
LOC100132354 significantly enhanced angiogenic capacity in the LAD cell lines through VEGFA/VEGFR2 signaling pathway and downstream Ras/Raf/Mek/Erk pathway
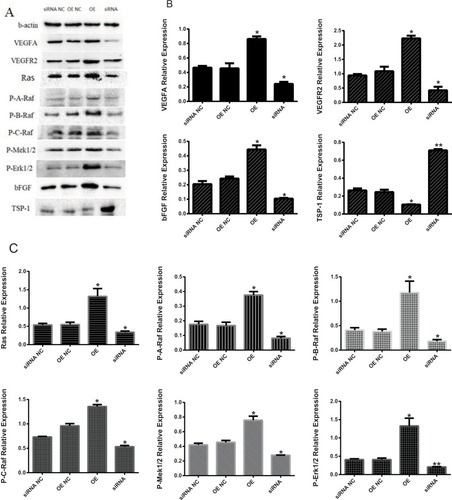
We further confirmed that the protein expression levels of VEGF, VEGFR2, bFGF, Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2 (t=7.258, 8.850, 8.994, 4.920, 9.032, 7.734, 9.141, 7.143; P=0.018, 0.013, 0.012, 0.039, 0.012, 0.024, 0.012, 0.019) were significantly higher in the OE group than in the OE NC group, while TSP-1 protein expression levels were lower (t=−7.471, P=0.017). The protein expression levels of VEGF, VEGFR2, bFGF, Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2 (t=8.701, 7.574, 6.491, 5.421, 5.578, 5.443, 10.140, 8.424, 12.689; P=0.013, 0.021, 0.023, 0.032, 0.031, 0.032, 0.010, 0.014, 0.006) in the siRNA group were lower than those of the siRNA NC group, but the TSP-1 protein expression levels were higher than those of the siRNA NC group (t=−24.005, P=0.002) ().
Figure 3 The proteins of VEGFA, VEGFR2, bFGF, TSP-1, Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2 from different cell lines by Western blotting.
Notes: (A) The Western blotting figure of these proteins. (B) The results of VEGFA, VEGFR2, bFGF, TSP-1 in the four groups. (C) The results of Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2 in the four groups. *P<0.05, **P<0.01.
Abbreviations: bFGF, basic fibroblast growth factor; OE, overexpression; NC, negative control; TSP-1, thrombospondin-1; VEGFA, vascular endothelial growth factor A; VEGFR2, vascular endothelial growth factor receptor-2.
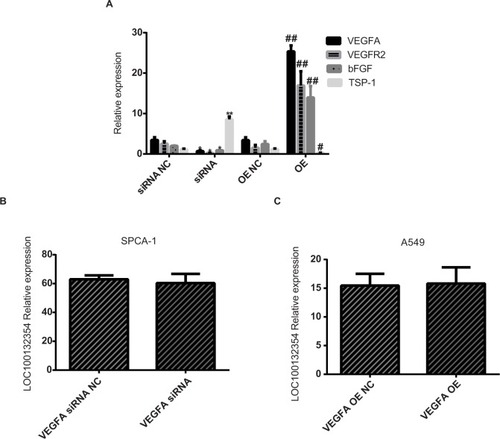
Compared with the OE NC group, the expression levels of VEGFA, VEGFR2, and bFGF mRNA (t=15.796, 16.041, 15.578; P=0.003, 0.002, 0.003) in the OE group were increased, while TSP-1 mRNA level was decreased (t=6.244; P=0.025). Compared with the siRNA NC group, the expression levels of VEGFA, VEGFR2, and bFGF mRNA (t=5.399, 5.903, 5.556; P=0.032, 0.023, 0.036) were decreased in the siRNA group, while TSP-1 mRNA level was increased (t=14.477; P=0.005) ().
Figure 4 The expression levels of these genes in each treatment group.
Notes: (A) Compared with the OE NC group, the expression level of VEGFA, VEGFR2, and bFGF mRNA (t=15.796, 16.041, 15.578; P=0.003, 0.002, 0.003) are higher in the OE group, while TSP-1 mRNA level was decreased (t=6.244; P=0.025). Compared with the siRNA NC group, the expression level of VEGFA, VEGFR2, and bFGF mRNA (t=5.399, 5.903, 5.556; P=0.032, 0.023, 0.036) are lower in the siRNA group, while TSP-1 mRNA level was increased (t=14.477; P=0.005). (B) LOC100132354 RNA expression level did not change after VEGFA knockdown (t=0.538; P=0.644). (C) LOC100132354 RNA expression level did not change after VEGFA overexpression (t=0.144; P=0.898). *P<0.05,**P<0.01, #P<0.05, ##P<0.01.
Abbreviations: bFGF, basic fibroblast growth factor; OE, overexpression; NC, negative control; TSP-1, thrombospondin-1; VEGFA, vascular endothelial growth factor A; VEGFR2, vascular endothelial growth factor receptor-2.
These results confirmed that after inhibition or overexpression of LOC100132354, the key molecular proteins and mRNA levels in the VEGFA/VEGFR2 signaling pathway of the cell line were significantly changed.
VEGFA is a downstream target gene of LOC100132354
LOC100132354 RNA expression level showed no obvious changes after VEGFA knockdown (t=0.538; P=0.644) or overexpression (t=0.144; P=0.898; ), suggesting that VEGFA was a downstream target gene of LOC100132354.
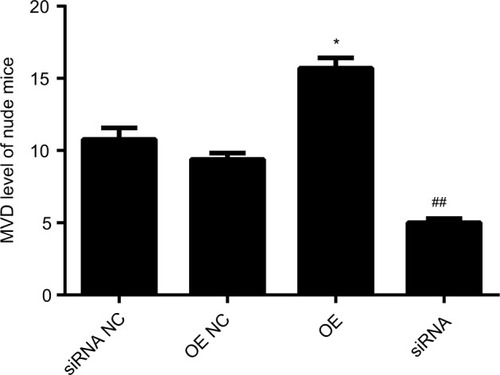
LOC100132354 significantly enhanced the MVD value of the nude mice
Our previous work showed that LOC100132354 significantly enhanced the tumorigenic ability in the nude mice experiment.Citation14 Tumorigenesis of nude mice confirmed that the tumor MVD was decreased significantly (t=10.765; P=0.012) after inhibition of LOC100132354 and increased significantly (t=15.879; P=0.009) after overexpression of LOC100 132,354 ( and ).
Table 3 Comparison of MVD in nude mouse tumors (x±SD) (n=25)
Figure 5 Changes of MVD expression in tumors of nude mice.
Notes: The tumorigenesis of nude mice confirmed that after inhibition of LOC100132354, the tumor MVD was decreased significantly (t=10.765; P=0.012) and after overexpression of LOC100 132354, the MVD was increased significantly (t=15.879; P=0.009). *P<0.05, ##P<0.01.
Abbreviations: MVD, microvessel density; OE, overexpression; NC, negative control.
Discussion
Tumor cells secrete VEGFA and bind to the VEGFR2 on the surface of endothelial cells to exert their biological effects.Citation17,Citation18,Citation34 Studies have shown that the expression levels of VEGFA and VEGFR2 were significantly elevated in NSCLC tissues,Citation35,Citation36 indicating that angiogenesis was significantly enhanced in NSCLC tumors. Steady expression of VEGFA can lead to increase in tumor volume in lung cancer in mice,Citation36 while inhibition of VEGFA signaling pathways resulted in the inhibition of tumor growth and tumor angiogenesis.Citation37,Citation38
In this study, the mRNA expression levels of LOC100132354, VEGFA, VEGFR2, and bFGF were significantly higher than the corresponding NT tissues, while TSP-1 mRNA are significantly lower than the corresponding NT tissues in LAD. The survival analysis showed that VEGFA, VEGFR2, and bFGF were poor predictors, while TSP-1 was a good predictor in LAD. These results revealed that the angiogenic capacity was enhanced significantly and could predict the prognosis of LAD.
In our previous studies, we found that LOC100132354 increased the ability of colony formation, cell migration, adhesion, proliferation, invasion, S phase, and G2/M phase cells, while decreased G0/G1 phase cells and apoptosis.Citation14 In this study, we further confirmed that the expression levels of VEGFA, VEGFR2, bFGF, Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2 were significantly higher from the OE group than those of the OE NC group, while TSP-1 expression levels were decreased. This showed that over-expression of LOC100132354 activated VEGFA/VEGFR2 pathway, further activating the downstream molecular Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2. Otherwise, VEGFA raised the levels of bFGF, while vascular growth negative regulator-TSP-1 protein expression levels were decreased. The expression levels of VEGFA, VEGFR2, bFGF, Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2 in the siRNA group were lower than those of the siRNA NC group, and only TSP-1 expression level was higher than that of the siRNA NC group. This suggested that the knockdown of LOC100132354 inactivated VEGFA/VEGFR2 pathway, further inactivating the downstream molecular Ras, P-A-Raf, P-B-Raf, P-C-Raf, P-Mekl/2, and P-Erk1/2. Otherwise, VEGFA decreased the levels of bFGF, while TSP-1 expression levels were increased. So, the knockdown and overexpression experiments consistently confirmed that LOC100132354 regulated the angiogenic capacity by VEGFA/VEGFR2 pathways in LAD.
LOC100132354 RNA expression levels showed no changes after VEGFA knockdown or overexpression, otherwise LOC100132354 could significantly enhance the MVD value of the nude mice. This suggested that VEGFA was a downstream target gene of LOC100132354.
Conclusion
Our study ascertained that LOC100132354 increased the angiogenic capacity and the mechanism of regulation of angiogenic capacity in LAD. Our results confirmed that lncRNA LOC100132354 can act as an antiangiogenic target in LAD.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (81672088), Zhejiang Provincial Natural Science Foundation (LQ16H160020), Zhejiang Provincial Health Planning Commission (2018KY514), and the Wenzhou Municipal Science and Technology Bureau of China (Y20170208, Y20170718).
Disclosure
The authors report no conflicts of interest in this work. There are not any of the family relationship between such authors.
References
- JemalAMurrayTWardECancer statistics, 2005CA Cancer J Clin2005551103015661684
- GridelliCRossiAMaionePTreatment of non-small-cell lung cancer: state of the art and development of new biologic agentsOncogene200322426629663814528288
- StewartDJTumor and host factors that may limit efficacy of chemotherapy in non-small cell and small cell lung cancerCrit Rev Oncol Hematol201075317323420047843
- ChenCHLaiJMChouTYVEGFA upregulates FLJ10540 and modulates migration and invasion of lung cancer via PI3K/AKT pathwayPLoS One200944e505219337377
- OgawaETakenakaKKatakuraHPerimembrane Aurora-A expression is a significant prognostic factor in correlation with proliferative activity in non-small-cell lung cancer (NSCLC)Ann Surg Oncol200815254755418043979
- RachetBWoodsLMMitryECancer survival in England and Wales at the end of the 20th centuryBr J Cancer200899Suppl 1S2S1018813248
- ChenWZhengRBaadePDCancer statistics in China, 2015CA Cancer J Clin201666211513226808342
- JiangCYangYYangYLong noncoding RNA (lncRNA) HOTAIR affects tumorigenesis and metastasis of non-small cell lung cancer by upregulating miR-613Oncol Res201826572573429187267
- WangQChengNLiXCorrelation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinomaOncotarget2017822558256727911863
- ChenWZhaoWZhangLMALAT1-miR-101-SOX9 feedback loop modulates the chemo-resistance of lung cancer cell to DDP via Wnt signaling pathwayOncotarget2017855943179432929212230
- ThaiPStattSChenCHLiangECampbellCWuRCharacterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell linesAm J Respir Cell Mol Biol201349220421123672216
- YangYLiHHouSHuBLiuJWangJThe noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cellPLoS One201385e6530923741487
- HanLKongRYinDDLow expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLCMed Oncol201330469423979857
- ChenJZhangFWangJLncRNA LINC01512 promotes the progression and enhances oncogenic ability of lung adenocarcinomaJ Cell Biochem2017118103102311028569418
- ZhangLZhouXFPanGFZhaoJPEnhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinomaBiomed Pharmacother201468440140724721325
- WeisSMChereshDATumor angiogenesis: molecular pathways and therapeutic targetsNat Med201117111359137022064426
- CarmelietPJainRKMolecular mechanisms and clinical applications of angiogenesisNature2011473734729830721593862
- SakuraiTKudoMSignaling pathways governing tumor angiogenesisOncology201181Suppl 1242922212932
- KuhnertFKirshnerJRThurstonGDll4-Notch signaling as a therapeutic target in tumor angiogenesisVasc Cell2011312021923938
- FerraraNGerberHPLecouterJThe biology of VEGF and its receptorsNat Med20039666967612778165
- RobinsonCJStringerSEThe splice variants of vascular endothelial growth factor (VEGF) and their receptorsJ Cell Sci2001114Pt 585386511181169
- ChatterjeeSHeukampLCSiobalMTumor VEGF:VEGFR2 autocrine feed-forward loop triggers angiogenesis in lung cancerJ Clin Invest201312341732174023454747
- HamerlikPLathiaJDRasmussenRAutocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growthJ Exp Med2012209350752022393126
- LuKVChangJPParachoniakCAVEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complexCancer Cell2012221213522789536
- TakahashiSVascular endothelial growth factor (VEGF), VEGF receptors and their inhibitors for antiangiogenic tumor therapyBiol Pharm Bull201134121785178822130231
- VlodavskyIKornerGIshai-MichaeliRBashkinPBar-ShavitRFuksZExtracellular matrix-resident growth factors and enzymes: possible involvement in tumor metastasis and angiogenesisCancer Metastasis Rev1990932032261705486
- LawlerJThrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growthJ Cell Mol Med20026111212003665
- GlikiGWheeler-JonesCZacharyIVascular endothelial growth factor induces protein kinase C (PKC)-dependent Akt/PKB activation and phosphatidylinositol 3′-kinase-mediates PKC delta phosphorylation: role of PKC in angiogenesisCell Biol Int200226975175912377207
- RenSPengZMaoJHRNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicingsCell Res201222580682122349460
- MoindrotBCeraseACokerHA pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencingCell Rep201512456257226190105
- GongCMaquatLElncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elementsNature2011470733328428821307942
- WeidnerNFolkmanJPozzaFTumor angiogenesis: a new significant and independent prognostic indicator in early-stage breast carcinomaJ Natl Cancer Inst19928424187518871281237
- MaedaKChungYSOgawaYPrognostic value of vascular endothelial growth factor expression in gastric carcinomaCancer19967758588638608475
- LohelaMBryMTammelaTAlitaloKVEGFs and receptors involved in angiogenesis versus lymphangiogenesisCurr Opin Cell Biol200921215416519230644
- BonnesenBPappotHHolmstavJSkovBGVascular endothelial growth factor A and vascular endothelial growth factor receptor 2 expression in non-small cell lung cancer patients: relation to prognosisLung Cancer200966331431819324448
- MaYPYangYZhangSEfficient inhibition of lung cancer in murine model by plasmid-encoding VEGF short hairpin RNA in combination with low-dose DDPJ Exp Clin Cancer Res2010295620497582
- DeaconKOnionDKumariRWatsonSAKnoxAJElevated SP-1 transcription factor expression and activity drives basal and hypoxia-induced vascular endothelial growth factor (VEGF) expression in non-small cell lung cancerJ Biol Chem201228747399673998122992725
- LiXWangXYeHPengAChenLBarbigerone, an isoflavone, inhibits tumor angiogenesis and human non-small-cell lung cancer xenografts growth through VEGFR2 signaling pathwaysCancer Chemother Pharmacol201270342543722814678
