Abstract
Objective
microRNAs are regulatory molecules regarded as important in the pathogenesis of different types of tumors. microRNA-216a (miR-216a-5p) has been identified as a tumor suppressor in multiple malignancies. However, the role of miR-216a-5p in the pathogenesis of small cell lung cancer (SCLC) remains obscure. The objective of this study was to investigate the role of the miR-216a-5p/Bcl-2 axis in SCLC pathogenesis.
Materials and methods
All the experimental methods used were as follows: microarray analysis, cell culture, transient, and stable gene transfection; real-time fluorescence PCR; Western blot; flow cytometry for cell cycle analysis; in vitro proliferation assay; in vitro wound healing experiment; in vivo xenograft model in nude mice; and dual luciferase reporter assay. All statistical analyses were carried out using GraphPad Prism 7 software. Statistical significance was analyzed by Student’s t-test or one-way ANOVA. P <0.05 (typically compared with the negative control group) was considered as significant and is marked with an asterisk in the figures.
Results
In this study, we observed that miR-216a-5p is downregulated in SCLC cell lines compared to that in the normal human bronchial epithelial cell line 16-HBE. In vitro and in vivo experiments demonstrate that upregulation of miR-216a-5p significantly decreased cell growth and migration and its downregulation increased SCLC cell proliferation and migration and influenced the cell cycle. Using bioinformatics analyses, we predicted that the important antiapoptotic gene Bcl-2 is targeted by miR-216a-5p, and we identified a functional miR-216a-5p binding site in the 3′-UTR of Bcl-2 using luciferase reporter assay. Furthermore, we determined that suppression of miR-216a-5p modulated the expression of Bcl-2, Bax, and Bad proteins (Bcl-2 family proteins), while Bcl-2 knockdown abrogated the effect of miR-216a-5p downregulation on cell proliferation, cell migration, and the cell cycle.
Conclusion
Taken together, these findings suggest that miR-216a-5p regulates SCLC biology via Bcl-2 family proteins. Therefore, our study highlights the role of the miR-216a-5p/Bcl-2 axis in SCLC pathogenesis.
Keywords:
Introduction
Small cell lung cancer (SCLC) accounts for ~15% of all lung cancer cases.Citation1 SCLC tumors generally become chemoresistant after effective chemotherapy treatments, and most patients die from a recurrence of cancer or from serious complications.Citation2 Hence, it is important to explore the molecular mechanisms underlying SCLC progression. microR-NAs (miRNAs), a type of small noncoding RNA with a length of 19–25 nucleotides, can inhibit the expression of their target genes by binding to their 3′-UTR, resulting in mRNA degradation or posttranslational repression.Citation3 miRNAs are involved in the differentiation of normal tissues and play an important role in the pathogenesis of many human cancers.Citation3 Some previous studies have demonstrated that miRNAs are deregulated in SCLC progression.Citation1,Citation4–Citation6 According to our previous findings, miR-216a-5p was identified among 24 miRNAs that were abnormally expressed in SCLC H69/H69AR cells based on TaqMan miRNA micro-array analysis.Citation7,Citation8 A number of studies have demonstrated that miR-216a-5p is downregulated in gastric cancer, pancreatic cancer, and colorectal cancer and acts as a tumor suppressor.Citation9–Citation12 Therefore, experiments to determine whether miR-216a-5p acts as an oncogene or a tumor suppressor in SCLC are warranted.
Materials and methods
Microarray analysis
For miRNA microarrays (LC Sciences, Houston, TX, USA), differentially expressed miRNAs with a fourfold decrease in the raw expression level were ordered by P-value. The experimental procedures were performed as previously described in detail.Citation7
Cell culture
The human SCLC cell lines NCI-H69, NCI-H69AR, NCI-H446, and 16-HBE cells were all from the American Type Culture Collection (ATCC; Manassas, VA, USA), maintained at 37°C in a 5% CO2 humidified incubator and grown in Roswell Park Memorial Institute (RPMI)-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (HyClone, Logan, UT, USA). The H69AR cell line is a multidrug-resistant cell line that is primarily used in microarrays to screen for chemoresistance-related miRNAs in SCLC and is occasionally used in this study.Citation7
Transient and stable transfection
The siRNAs, miR-216a-5p mimics, inhibitors, and their respective negative control (NC) vectors were purchased from GenePharma (Shanghai, China) and then transiently and stably transfected according to the manufacturer’s instructions. The most effective interference sequences within Bcl-2 were identified based on real-time quantitative PCR (RT-qPCR) and then used for subsequent in vitro experiments. The stable overexpression or silence miR-216a-5p expression cell lines and control cell lines were constructed by lentivirus infection. The detailed procedure is provided in the “Supplementary materials” section. The relevant mimic and inhibitor sequences are listed in the “Supplementary materials” section.
RT-qPCR
RT-qPCR was used to detect the expression level of miR-216a-5p and genes in SCLC cells according to the manufacturer’s instructions (Takara, Dalian, China). GAPDH was used as the control. The relevant primers are listed in the “Supplementary materials” section.
Western blot analysis
Western blot analysis was performed according to standard protocols as previously described.Citation13 GAPDH and β-actin were used as the controls. The antibody information for apoptosis-related genes is listed in the “Supplementary materials” section.
Flow cytometry for cell cycle analysis
Cell cycle assays were performed after cells were fixed in 70% ethanol overnight at 4°C and then stained with propidium iodide. The procedure was conducted according to standard protocols described in detail previously.Citation15
In vitro proliferation assay
The Cell Counting Kit-8 (CCK-8; Dojindo, Kyushu Island, Japan) was used for the in vitro proliferation assays in accordance with the manufacturer’s instructions as described previously.Citation14,Citation15
In vitro wound healing experiment
SCLC cells were grown to confluence on 24-well plates. Consistently shaped wounds were made using a sterile 10 µL pipette tip. At least four images of the scraped area were captured using phase contrast microscopy immediately after the scratch and after 0, 12, 24, 36, 48, and 72 hours. The procedure was replicated three times and was performed as previously described.Citation16 The same scratched area was selected for measurement at each time point.
In vivo xenograft model in nude mice
Six-to-seven-week-old female BALB/c mice (Guangdong Experimental Animal Center, Guangzhou, China) were accommodated (license number for laboratory animals: SYXY (YUE)2013-0002) and handled in compliance with the current European, national, and local (RD 1201/2005) regulations, following the Federation for Laboratory Animal Science Associations and Animal Research: Reporting of In Vivo Experiments guidelines and with the approval of the Institutional Animal Care Use Committee of Sun Yat-Sen University.
The xenograft tumor formation experiment was conducted according to our standard protocol as described previously.Citation16 Animals were randomly divided into four groups (n=3 mice per group): 1) H69AR/LV-miR-NC; 2) H69/LV-miR-216a-5p inhibitors; 3) H69AR/LV-miR-216a-5p mimics; and 4) H69/LV-miR-NC. When the average longest tumor diameter of any group reached ~1 cm, the mice were sacrificed, and the excised tumors were measured.
Dual luciferase reporter assay
H69 cells were transfected with hsa-miR-216a-5p mimics/inhibitors/NC by Lipofectamine 2000 (Thermo Fisher Scientific). Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to check the luciferase activity of H69 cell lysates. The detailed procedure was conducted according to our standard protocol as described previously.Citation17 All treatments were performed in triplicate.
Statistical analyses
All experiments were conducted in triplicate. The data are expressed as mean ± SD. All statistical analyses were carried out using GraphPad Prism 7 software. Statistical significance was analyzed by Student’s t-test or one-way ANOVA. P<0.05 (typically compared with the NC group) was considered as significant and is marked with an asterisk in the figures.
Results
Expression of miR-216a-5p was determined in SCLC cell lines
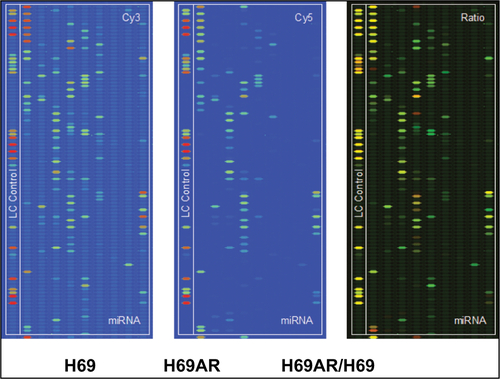
To identify miRNAs differentially expressed in SCLC, we performed TaqMan miRNA microarray analysis on the H69 and H69AR cell lines (ATCC; Supplementary materials). In all, 24 miRNAs with statistically significant differences in expression of more than fourfold were identified ( and ). Our previous studies have demonstrated that Bcl-2 had been proved to be involved in the development of SCLC, even in the occurrence of drug resistance.Citation18 We also found that Bcl-2 contains putative regions that match to the seed sequence of miR-216a-5p but not to other 23 miRNAs by further searching in RNA22-seq database (data not shown). Therefore, miR-216a-5p was chosen as an miRNA of interest for further study. By further verification with RT-qPCR, miR-216a-5p showed a significant downregulation in SCLC cell lines compared with that in the normal human bronchial epithelial cell line 16-HBE (ATCC), and the expression of miR-216a-5p in H69 cells was higher than that in H446 cells (ATCC; ). Accordingly, the H69 cell line, which exhibits a high level of expression of miR-216a-5p, was used for loss-of-function experiments, and the H446 cell line was used for gain-of-function experiments.
Table 1 Partial differential expression of miRNAs in SCLC H69/H69AR cells
Figure 1 Relative expression of miR-216a-5p in cell lines.
Notes: (A) Differentially expressed transcripts of miRNAs are provided following the chip images. (B) Expression level of miR-216a-5p in three SCLC cell lines and the 16-HBE cell line shows downregulation in SCLC cell lines, especially in the H69AR cell line, compared with that in the 16-HBE cell line. (C) Identification of miR-216a-5p upregulation in H69 cells and downregulation in H446 cells. For stable miR-216a-5p upregulation and downregulation, the inhibitors and mimics of miR-216a-5p were packaged into a lentivirus for subsequent studies. The transfection efficacy was validated by fluorescence microscopy in H69 cells (D) and RT-qPCR in both the cell lines (E). *P<0.05 and **P<0.001.
Abbreviations: miRNA, microRNA; SCLC, small cell lung cancer; RT-qPCR, real-time quantitative PCR.
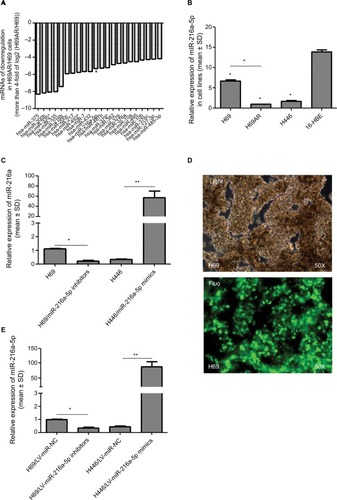
In order to manipulate miR-216a-5p levels in SCLC cells, miR-216a-5p mimics and inhibitors (GenePharma) were transfected into H446 and H69 cells, respectively. RT-qPCR analysis of miR-216a-5p expression was performed at 24 hours after transfection, and the results revealed that the miR-216a-5p level was effectively increased or decreased. The observed inhibition level of miR-216a-5p expression caused by miR-216a-5p inhibitor treatment was 79.7%, and its level of overexpression caused by miR-216a-5p mimic treatment was 168.04% (). The miR-216a-5p mimics and inhibitors were used in subsequent loss- and gain-of-function studies. For stable transfection, the mimics and inhibitors were packaged into a lentiviral vector for the subsequent studies (), and analysis demonstrated that their ability to increase or decrease expression of miR-216a-5p was significant ().
miR-216a-5p regulates SCLC cell growth in vitro and in vivo
To investigate the effect of miR-216a-5p on the pathogenesis of SCLC in vitro, a CCK-8 assay was carried out in miR-216a-5p up- and downregulated cells. The CCK-8 assay revealed that downregulation of miR-216a-5p increased H69 cell proliferation compared with that in the NC group, while upregulation of miR-216a-5p decreased cell proliferation ().
Figure 2 miR-216a-5p inhibits SCLC cell proliferation and migration in vitro and decreases growth of xenografts in nude mice.
Notes: (A) A CCK-8 assay was used to determine the effect of miR-216a-5p up- or downregulation on SCLC cell proliferation. (B) Images of excised tumors from tumor-bearing nude mice in each group. (C) Tumor volume change for each group measured on the indicated days. (D) Flow cytometric analysis was used for cell cycle phase detection in miR-216a-5p-downregulated H69 cells. (E) miR-216a-5p suppressed SCLC cell migration. (F) Quantitative results of the wound closure rate for miR-216a-5p-upregulated H446 cells. *P<0.05.
Abbreviations: SCLC, small cell lung cancer; CCK-8, Cell Counting Kit-8; NC, negative control.
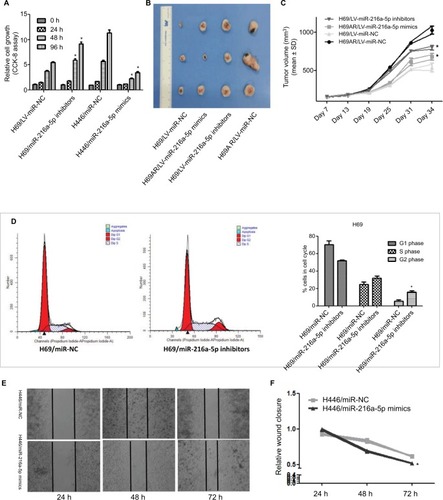
The ability of miR-216a-5p to confer SCLC characteristics was examined using an in vivo xenograft model in nude mice. As shown in , tumor growth in mice was significantly increased in the H69/LV-miR-216a-5p inhibitors group compared to that in the H69/LV-miR-NC group and was significantly decreased in the H69AR/LV-miR-216a-5p mimics group compared to that in the H69AR/LV-miR-NC group. At 21 days after subcutaneous injection, the mean tumor volume for the H69/LV-miR-216a-5p inhibitors group was markedly larger than that for the H69/LV-miR-NC group and that for the H69AR/LV-miR-216a-5p mimics group was smaller compared to that of the H69AR/LV-miR-NC group ().
miR-216a-5p suppression resulted in cell cycle disorder
To further investigate the growth promotion observed in miR-216a-5p downregulated cells, cell cycle profiles of miR-216a-5p downregulated cells were evaluated using flow cytometry. The suppression of miR-216a-5p led to cell blockade characterized by G2/M phase arrest and an increase in the number of SCLC cells in the G2/M phase ().
miR-216a-5p promotes cell migration in vitro
A scratch healing test was used to determine the effect of miR-216a-5p on cell migration. The results showed that miR-216a-5p upregulation led to a significant reduction in the wound closure rate in H446 cells ().
Bcl-2 is predicted and confirmed to be a miR-216a-5p target gene
Using miRNA target detection software RNA22-seq (https://cm.jefferson.edu/), miR-216a-5p was predicted to bind to the apoptosis-related gene Bcl-2 (). The predicted binding relationship was confirmed by RT-qPCR and Western blot in SCLC cells. The results demonstrated that miR-216a-5p acts as an inhibitor of Bcl-2 (). Moreover, inhibition of miR-216a-5p expression in H69 and H69AR cells led to a significantly corresponding increase in Bcl-2 expression ().
Figure 3 Bcl-2 is a target gene of miR-216a-5p.
Notes: (A) Putative binding site of miR-216a-5p in the Bcl-2 3′-UTR and the site of target mutagenesis. (B) miR-216a-5p negatively regulated the expression of its target gene Bcl-2. (C) miR-216a-5p negatively regulated the expression of the Bcl-2 protein. (D) Binding and mutation sequences used in the luciferase reporter assay. (E) The luciferase reporter assay was used to demonstrate the target binding relationship between miR-216a-5p and Bcl-2. *P<0.05.
Abbreviation: NC, negative control.
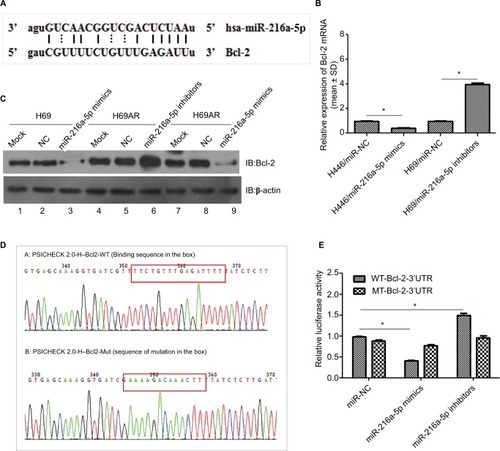
To further investigate the regulatory relationship between miR-216a-5p and Bcl-2, we constructed Bcl-2 wild-type and mutant psicheck-2.0 luciferase vectors and conducted a luciferase reporter assay (). Luciferase activity was reduced by ~59% () when miR-216a-5p was upregulated and was increased by 50.1% when miR-216a-5p was downregulated. These data demonstrate that miR-216a-5p binds directly to the Bcl-2 3′-UTR through its miRNA binding sites, leading to miRNA-induced repression of luciferase reporter activity.
miR-216a-5p regulates the expression of apoptosis-related gene Bcl-2/Bax/Bad
To investigate the regulatory effect of miR-216a-5p on its target gene Bcl-2, RT-qPCR was used to verify that down-regulation of miR-216a-5p leads to an increase in Bcl-2 mRNA expression. Additional experiments showed that siRNA-mediated Bcl-2 knockdown rescued the overexpression of Bcl-2 caused by miR-216a-5p inhibition (). Hence, a direct relationship between miR-216a-5p and Bcl-2 was confirmed.
Figure 4 Bcl-2 mediates the effect of miR-216a-5p on SCLC pathogenesis.
Notes: (A) Rescue experiment of Bcl-2 downregulation by miR-216a-5p knockdown using RT-qPCR. (B) and (C) miR-216a-5p mediated expression of the apoptosis-related genes Bcl-2/Bad/Bax at the mRNA and protein levels. (D) miR-216a-5p decreased cell growth by targeting Bcl-2. (E) and (F) miR-216a-5p influenced the cell cycle of SCLC cells by targeting Bcl-2. (G) The in vitro wound healing experiment shows that Bcl-2 knockdown rescued the effect of miR-216a-5p downregulation on SCLC cell migration. *P<0.05.
Abbreviations: SCLC, small cell lung cancer; RT-qPCR, real-time quantitative PCR; NC, negative control; CCK-8, Cell Counting Kit-8.
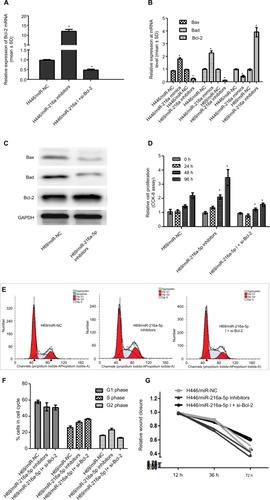
To determine whether miR-216a-5p-mediated regulation of the important apoptosis suppressor gene Bcl-2 is incidental, we performed RT-qPCR and Western blot analysis to assess the effect of miR-216a-5p on the expression of the apoptosis-promoting genes Bax and Bad. The results demonstrated that miR-216a-5p downregulation led to a significant decrease in the mRNA () and protein levels () of both Bax and Bad. Previous studies indicated that miR-216a-5p promotes the occurrence of apoptosis in SCLC. We speculated that miR-216a-5p participates in the development of SCLC by promoting apoptosis and affecting the apoptosis pathway.
Bcl-2 partly mediates the effect of miR-216a-5p on SCLC cell proliferation and migration
Interestingly, we found that Bcl-2 knockdown also caused a significant inhibition of SCLC cell growth, as measured by CCK-8 assay, just as miR-216a-5p upregulation did (). Moreover, flow cytometry experiment revealed that Bcl-2 knockdown rescued the impact of miR-216a-5p downregulation on cells arrested in the G2 phase, but the S-phase does not seem to be impacted by Bcl-2 siRNA (). Meanwhile, different from the increase in cell migration caused by miR-216a-5p downregulation, as shown in the wound healing assay, Bcl-2 knockdown led to a reduction in cell migration ability compared with the NC treatment (). Taken together, these results further substantiate the presumptive role of miR-216a-5p and Bcl-2 in SCLC cell cycle progression and provide new evidence to support the concept of their interdependent expression.
Discussion
SCLC is an aggressive malignancy characterized by high incidence, rapid development, and low survival rate.Citation19,Citation20 Although aberrant expression of miRNAs in lung cancer has been reported, few studies have elucidated the role of specific miRNAs in SCLC tumorigenesis.Citation21 In the present study, miRNA microarray analysis identified 24 miRNAs with a significantly downregulated expression in the SCLC che-moresistant H69AR cell line compared with that in the H69 cell line. Therefore, we focused on analyzing miR-216a-5p in detail. Our previously published microarray study, which screened SCLC lines for chemoresistance-associated miR-NAs,Citation7 surprisingly found that miR-216a-5p was expressed in low quantities based on RT-qPCR. Our study demonstrates that miR-216a-5p plays an important role in SCLC cell growth and migration and modulates the G2 phase of the cell cycle.
Bcl-2 was subsequently identified as a direct target gene of miR-216a-5p that is involved in SCLC pathogenesis under negative regulation by miR-216a-5p. In addition to its effect on Bcl-2, miR-216a-5p downregulation promoted the expression of the apoptosis-related genes Bax and Bad. Bcl-2, a well-known apoptosis suppressor gene, is involved in the progression of multiple tumors.Citation22,Citation23 Studies have reported that many miRNAs can regulate the expression of Bcl-2.Citation24 However, the mechanism through which Bcl-2 participates in SCLC biology under the regulation of miR-216a-5p remained obscure. Based on our previous study, Bcl-2 was found to play a key role in the chemoresistance of SCLC due to its antiapoptotic function.Citation18 The data in this study showed that Bcl-2 promotes cell proliferation and tumorigenesis in nude mice by modulating the cell cycle. Furthermore, when we downregulated Bcl-2 expression by cotransfection with Bcl-2 siRNA in a stable cell line in which miR-216a-5p was downregulated, the upregulation of Bcl-2 was reversed and cell proliferation was inhibited, which is consistent with the effect of miR-216a-5p upregulation on cell proliferation. These results further suggest that miR-216a-5p might regulate SCLC cell proliferation through mediation of Bcl-2 expression. Taken together, the results of this study indicate that miR-216a-5p may affect SCLC biology by activating the apoptosis signaling pathway, although more in-depth study will be needed.
Numerous studies have closely linked miR-216a-5p to a variety of human tumors. For example, miR-216a-5p was reported to be downregulated by more than 200-fold in pancreatic ductal adenocarcinoma samples compared to that in normal pancreatic tissues, while the same situation was not observed in chronic pancreatitis samples. Similarly, upon transformation of pancreatic stellate cells to myofibroblast-like cells,Citation18 a key step in pancreatic cancer progression, miR-216a-5p was downregulated. Moreover, the downregulation of miR-216a-5p was associated with pancreatic ductal adenocarcinoma cell migration, invasion, and epithelial-to-mesenchymal transition and pancreatic cancer progression. On the basis of those studies, our findings confirm that miR-216a-5p is a tumor suppressor.
In this study, we first validated our previous TaqMan miRNA microarray results by RT-qPCR and chose miR-216a-5p as the subject of our investigation of SCLC. Second, on the basis of up- and downregulation of miR-216a-5p expression through the use of miR-216a-5p mimics and inhibitors, respectively, we found in in vitro and in vivo experiments that miR-216a-5p inhibited cell proliferation, cell migration, and xenograft formation and influenced cell cycle status, particularly in the G2 phase. Finally, using bioinformatics and a luciferase reporter assay, we confirmed that miR-216a-5p targets the Bcl-2 gene. Bcl-2 promoted cell proliferation and migration and impacted the G2 phase of the cell cycle under regulation by miR-216a-5p. Furthermore, suppression of miR-216a-5p inhibited the expression of Bax and Bad while increasing Bcl-2 expression. Our study suggests that miR-216a-5p is involved in SCLC pathogenesis via modulation of Bcl-2/Bax/Bad.
Moreover, our study demonstrated that the suppression of proliferation capacity of H69 cells by Bcl-2 gene silencing might be due to a reduction of cells in the G2 phase of the cell cycle. Since the S phase does not seem to be impacted by Bcl-2 siRNA and small molecule inhibitors (eg, venetoclax) of Bcl-2 impact SCLC cell line viability,Citation25 how do Bcl-2 siRNA or Bcl-2 inhibitors impact SCLC cells’ survival in the context of miR-216a expression will be our next area of interest.
Conclusion
We have demonstrated that miR-216a-5p plays an important role in the pathogenesis of SCLC. miR-216a-5p expression significantly decreased cell proliferation and migration in SCLC through mediating the expression of Bcl-2/Bax/Bad proteins. Based on these findings, we propose that miR-216a-5p may be used as a therapeutic agent for SCLC.
Supplementary materials
Figure S1 miRNA profiles from Cy3 and Cy5 images of H69AR/H69 cells by TaqMan miRNA microarray analysis.
Abbreviation: miRNA, microRNA.
Primers
miR-216a-5p RT primer (5′–3′): CTCAACTGGTGTCGTGGA
miR-216a-5p forward primer (5′–3′): TGTCGCAAATCT CTGCAGG
miR-216a-5p reverse primer (5′–3′): CAGAGCAGGGTCCGAGGTA
Bcl-2 forward primer (5′–3′): GAACTGGGGGAGGATTGTGG
Bcl-2 reverse primer (5′–3′): CCGTACAGTTCCACAAAGGC
Bax forward primer (5′–3′): CCCGAGAGGTCTTTTTCCGAG
Bax reverse primer (5′–3′): CCAGCCCATGATGGTTCTGAT
Bad forward primer (5′–3′): CCCAGAGTTTGAGCCGAGTG
Bad reverse primer (5′–3′): CCCATCCCTTCGTCGTCCTGAPDH forward primer (5′–3′): GGGCTGCTTTTAACTCTGGAPDH reverse primer (5′–3′): TGGCAGGTTTTTCTAGACGG
miR-216a-5p mimics and inhibitor sequences
Hsa-miR-216a-5p mimic: UAAUCUCAGCUGGCAACUGUGAACAGUUGCCAGCUGAGAUUAUU Hsa-miR-216a-5p inhibitor: UCACAGUUGCCAGCUGAGAUUA
Antibodies information
Rabbit antihuman monoclonal antibody Bcl-2 (#5284; Santa Cruz Biotechnology, Santa Cruz, CA, USA)
Rabbit antihuman monoclonal antibody Bad (#9239; Santa Cruz Biotechnology)
Rabbit antihuman monoclonal antibody Bax (#2772; Santa Cruz Biotechnology)
The stable overexpression or silence miR-216a-5p expression cell lines and control cell lines were constructed by lentivirus infection, which were purchased from GenePharma (Shanghai, China). Cell transfection was carried out using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81372508, 81702285), the Guangdong Natural Science Foundation (Special Fund for Scientific and Technological Development; 2017A030313644), the Guangdong Medical Research Foundation (A2017330), the fund for Innovative Experiments for College Students in Guangdong Medical University (ZYDM021), and the College Students’ Innovation and Entrepreneurship Training Program (GDMU2017133).
Disclosure
The authors report no conflicts of interest in this work.
References
- duLSchagemanJJIrnovMicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCsJ Exp Clin Cancer Res20102917520624269
- VermaVChoiJISimone CB 2nd. Proton therapy for small cell lung cancerTransl Lung Cancer Res20187213414029876312
- TaucherVManggeHHaybaeckJNon-coding RNAs in pancreatic cancer: challenges and opportunities for clinical applicationCell Oncol2016394295318
- MizunoKMatakiHAraiTThe microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenesJ Hum Genet201762767167828275243
- LiuHYChenJPolymorphisms in miRNA binding site: new insight into small cell lung cancer susceptibilityActa Pharmacol Sin201132101191119221874042
- XiongFWuCChangJGenetic variation in an miRNA-1827 binding site in MYCL1 alters susceptibility to small-cell lung cancerCancer Res201171155175518121676885
- GuoLLiuYBaiYGene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysisEur J Cancer20104691692170220371173
- RoaWBrunetBGuoLIdentification of a new microRNA expression profile as a potential cancer screening toolClin Invest Med2010332124
- HouBJianZChenSExpression of miR-216a in pancreatic cancer and its clinical significanceNan Fang Yi Ke Da Xue Xue Bao201232111628163123174591
- ZhangDZhaoLShenQDown-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancerInt J Cancer2017140102298230928213952
- WangDLiYZhangCLiXYuJMiR-216a-3p inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5J Cell Biochem201811921755176628786533
- WuYZhangJZhengYMaCLiuXESunXmiR-216a-3p Inhibits the Proliferation, Migration, and Invasion of Human Gastric Cancer Cells via Targeting RUNX1 and Activating the NF-κB Signaling PathwayOncol Res201826115717128835317
- LiHLiTFanJmiR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathwayCell Death Differ201522121935194526206089
- SuYBaiMZhuLPPirh2 shRNA mediated by psiRNA-hH1 vector plasmid effectively inhibits the proliferation of lung carcinoma cells: in vitro and in vivo experimentsZhonghua Yi Xue Za Zhi200787171199120317686242
- ZhouHZhengLLuKDownregulation of Cohesin Loading Factor Nipped-B-Like Protein (NIPBL) Induces Cell Cycle Arrest, Apoptosis, and Autophagy of Breast Cancer Cell LinesMedical Science Monitor20072348174825
- SunYZengCGanSLncRNA HOTTIP-Mediated HOXA11 Expression Promotes Cell Growth, Migration and Inhibits Cell Apop-tosis in Breast CancerInt J Mol Sci2018192472
- YeMWeiTWangQTSPAN12 promotes chemoresistance and proliferation of SCLC under the regulation of miR-495Biochem Biophys Res Commun2017486234935628302484
- SunYHuBWangQLong non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216aCell Death Dis2018928529367594
- BaiYWangYLYaoWJExpression of miR-32 in human non-small cell lung cancer and its correlation with tumor progression and patient survivalInt J Clin Exp Pathol20158182482925755781
- LiXXLiRJZhaoLJLiuNBWangPExpression of molecular factors correlated with metastasis in small cell lung cancer and their significanceInt J Clin Exp Pathol2015811146761468426823791
- PakMGLeeCHLeeWJShinDHRohMSUnique microRNAs in lung adenocarcinoma groups according to major TKI sensitive EGFR mutation statusDiagn Pathol2015109926170125
- TorrealbaNRodriguez-BerrigueteGFraileBPI3K pathway and Bcl-2 family. Clinicopathological features in prostate cancerAging Male2018112
- ReedJCBcl-2 on the brink of breakthroughs in cancer treatmentCell Death Differ20182513629227986
- SunXXZhangSSDaiCYLukS-PV-Regulated MicroRNA-125a-3p Promotes THP-1 Macrophages Differentiation and Apoptosis by Down-Regulating NF1 and Bcl-2Cell Physiol Biochem20174431093110529179212
- LochmannTLFlorosKVNaseriMVenetoclax is effective in small-cell lung cancer with high BCL-2 expressionClin Cancer Res201824236036929118061
