Abstract
Objective
A greater knowledge of the mechanisms of the pathogenesis of penile cancers may assist in the development of more tailored targeted therapy. Herein, we aimed to evaluate the expression of CEACAM19 in penile cancer and to explore its regulatory mechanisms.
Material and methods
This retrospective study enrolled 64 penile cancer patients who underwent penectomy between 2011 and 2015. CEACAM19 expression in tissues was detected by immunohistochemistry, which was analyzed in association with clinicopathological parameters. Kaplan–Meier analysis was performed to evaluate the relationship between CEACAM19 expression and prognosis of patients with penile cancer. Cell Counting Kit-8 assay and clonogenic assay were used to evaluate the cell viability and tumorigenic potential of penile cancer cell line, respectively; wound healing assay and transwell invasion assay were conducted to evaluate the effect of CEACAM19 depletion on cell migration and invasion in penile cancer cells; CEACAM19 protein expression was analyzed by Western blotting. Culture supranatant matrix metalloproteinase 2/9 (MMP2/9) was detected by ELISA.
Results
CEACAM19 was differentially expressed in non-cancerous and penile cancer tissues. Over-expression of CEACAM19 was significantly associated with nodal and distant metastasis, and predicted unfavorable cancer-specific survival in penile cancer. Depletion of CEACAM19 expression suppressed cell proliferation, reduced colony formation, and attenuated cell migration and invasion in Penl1 cells. Furthermore, knockdown of CEACAM19 expression attenuated the levels of p-Smad2/3 and reduced secretion of MMP2/9 in Penl1 cells. The effects of CEACAM19 might result from its function in regulating the Smad2/3 activation, as inhibition on Smad2/3 activation suppressed cell migration and invasion and reduced MMP2/9 secretion in Penl1 cells.
Conclusion
Over-expression of CEACAM19 might serve as a potential prognostic biomarker for clinical management of penile cancer. Strategies targeting CEACAM19-regulated signaling pathways may have a therapeutic benefit in penile cancer.
Keywords:
Introduction
Penile cancer, a relatively rare cancer, arises from the squamous epithelium of the glans, coronal sulcus, or inner preputial surface.Citation1 Penile squamous cell carcinoma comprises the majority of cases of penile cancer.Citation2 Currently, surgical operation is recommended as the major treatment for penile cancer. Besides the surgical approach, brachytherapy, chemotherapy, and targeted therapy were also applied in the clinical management of penile cancer.Citation3 Clinical outcomes of penile cancer are associated with tumor grade, pathological subtype, human papilloma virus (HPV) status, and clinical stage, with the presence of inguinal nodal metastases being the most important prognostic factor for cancer-specific survival (CSS).Citation4 Despite considerable progresses in clinical treatment, the survival of the patients with penile cancer has not improved during the previous 20 years.Citation5
Biomarkers are increasingly used in cancer diagnosis and clinical management. Improved understanding of the molecular/genetic mechanisms relating to the development of penile cancer has allowed for the clinical use of newer biomarkers.Citation6 Some molecular markers have already been studied in penile cancer, including HPV, p53, p16, SCC antigen, Ki-67, and Wnt/β-catenin.Citation6–Citation9 These markers are not good predictors of tumor progression/metastasis. The gene carcinoembryonic antigen related cell adhesion molecule 19 (CEACAM19) belongs to the CEACAM subfamily of the carcinoembryonic antigen family. The CEACAM19 gene is mapped to human chromosome 19q13.2 and is made up of eight exons interspaced with seven introns. The encoded protein contains two immunoglobulin-like transmembrane domains and a conserved motif of eukaryotic translation initiation factors (10). CEACAM19 is found to be mainly expressed in the prostate, uterus, fetal brain, mammary, adrenal gland, skeletal muscle, small intestine, and kidney.Citation10 Dysregulated CEACAM19 expression was observed in breast cancer and may serve as a biomarker for breast cancer progression.Citation11,Citation12 Herein, we sought to examine the expression of CEACAM19 in penile cancer and to determine its association with clinicopathological features. Our findings suggested that over-expression CEACAM19 was significantly associated with nodal status and distant metastasis in penile cancer. Therefore, CEACAM19 might be potentially useful as a novel biomarker or therapeutic target for penile cancer.
Materials and methods
Patient cohort
The patients enrolled in this study (n=64) were performed penectomy and diagnosed with penile cancer (2011–2015) in Xiangya Hospital Central South University. Patients with known chemotherapy or brachytherapy before the surgery were excluded from the study. The research protocols were approved by the institutional research ethics committee in Xiangya Hospital with written informed consent acquired from all the patients (No. 201805847). This study was conducted in accordance with the Declaration of Helsinki. The cases were reviewed by two genitourinary pathologists (YW and SW). Representative paraffin-embedded archival blocks were used for sectioning (4 µm thickness). All patients were prescribed a follow-up regimen based on the National Comprehensive Cancer Network guidelines, with physical examination every 6 months. Cancer and vital status were recorded by clinical follow-up at our department (median follow-up: 50.5 months). TNM staging was assigned based on the American Join Committee on Cancer, seventh edition.
Reagents and cell line
Primary antibodies are obtained from the following sources: CEACAM19 (HPA052865, Sigma, St. Louis, MO, USA); p-Smad2/3, Smad2/3, and β-actin (Cell Signaling, Beverly, MA, USA). Human penile cancer cell line Penl1 was kindly provided by Prof Hui Han (Department of Urology, Cancer hospital, Sun Yat-sen University).Citation13 The use of Penl1 in this study was approved by the institutional research ethics committee in Xiangya Hospital, Central South University. Penl1 was routinely cultured in DMEI supplemented with 10% FBS (Hyclone, Logan, UT, USA), 4 mM glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin. Lentiviral plasmids expressing scramble (Scr) or shCEACAM19 was purchased from Genecopoeia Inc. (Rockville, MD, USA). The packaging procedure for lentiviral shRNAs was conducted as described previously.Citation14
Immunohistochemistry (IHC)
IHC on tissue sections were performed as described previously.Citation14 Briefly, the sections were dewaxed in xylene, rehydrated with graded alcohols, and subjected to heat-induced epitope retrieval. The sections were then incubated overnight at 4°C with a rabbit polyclonal anti-CEACAM19 antibody (dilution, 1:50), followed by incubation with a horseradish peroxidase conjugated goat anti-rabbit secondary antibody (DAKO Denmark A/S, Glostrup, Denmark) for 30 minutes at room temperature. Immunochemical staining were visualized by reaction with 3,3′-diaminobenzidine and hydrogen peroxide chromogen substrate (DAKO). Slides were counterstained with hematoxylin and mounted with coverslip. The negative controls were incubated with isotype rabbit IgG. CEACAM19 expression was scored by two clinical pathologists (YW and SW). Immunostaining of CEACAM19 was assessed using a scoring system as described previously.Citation15 The staining positivity (≥30%) was regarded as over-expression.
HPV detection
HPV detection was conducted by SPF10 polymerase chain reaction and DEIA as described previously.Citation16 Briefly, total genomic DNA was extracted from FFPE tissue material using Qiagen QIAamp DNA Micro Kit. Each DNA isolation run and PCR run contained HPV positive and negative controls. Specimens were tested for HPV DNA by PCR amplification/typing using the HPV SPF10 PCR DEIA assay (Labo Biomedical Products, Rijswijk, Netherlands).
Clonogenic survival assay
Clonogenic survival assay was used to measure the tumorigenic potential of Penl1 cells.Citation17 Briefly, Penn1 cells (5×102) were plated in 6 cm culture dishes, and cultured for 12 days, and the number of surviving colonies (defined as a colony with >50 cells) was stained with 0.5% crystal violet and counted.
Cell viability analysis
Cell viability was determined by Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories, Kumamoto, Japan) as described previously.Citation18 Briefly, Penl1 cell were seeded (2×103 cells/well) into 96-well plates. After different time point in culture, CCK-8 solution (10 µL) was added and incubated for 1 hour at 37°C. The OD value (absorbance) was measured at 450 nm by Infinite F50 microplate reader (Tecan Group AG, Männedorf, Switzerland).
Western blot
Western blot was conducted as described previously.Citation19 Briefly, cells were lysed and quantified using Bicinchoninic Acid Protein Assay Kit (Beyotime Biotechnology, Shanghai, P.R. China). Protein lysates (15 µg) were separated by 10% SDS-PAGE and transferred to Millipore PVDF membranes. Blots were blocked with 5% non-fat dry milk in Tris-buffered saline buffer for 2 hours at room temperature and then incubated with diluted antibodies overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated secondary antibody (Abcam, Cambridge, MA, USA) for 1 hour at room temperature. The blotting signal was visualized using an enhanced chemiluminescence detection reagent (Abcam). β-Actin served as a loading control.
Wound healing assay
Wound healing assay was used to evaluate the cell migration ability as described previously.Citation20,Citation21 Briefly, Penl1 cells (5×105) were grown in six-well plates for 48 hours until the cells grew confluent. A sterile 200 µL pipette tip was used to make a scratch wounds in the center of the well. After 24 hours, the distance between the wound sides was measured, respectively.
Transwell invasion assay
Cell invasion assay was performed based on transwell chamber with 8 µm pores (Corning, Corning, NY, USA) as described previously.Citation22,Citation23 Briefly, Penl1 cells (5×105 cells/well) were seeded in transwell inserts pre-coated with 50 µL Matrigel (BD Biosciences, San Jose, CA, USA). The plates were incubated for 36 hours at 37°C. The invaded cells on the bottom surface of the membrane were fixed by dehydrated alcohol, and stained by 0.2% crystal violet solution (Sigma-Aldrich Co., St Louis, MO, USA). After wash with deionized water, Penl1 cells were photographed with Olympus BX43 microscope. The crystal violet in stained cells was eluted by 20% glacial acetic acid and measured with Infinite F50 microplate reader (Tecan, Switzerland) at 570 nm.
Luciferase reporter assay on cancer-related signaling pathway
Analysis on cancer-related pathways activity was performed using the Cignal Finder Pathway Reporter Arrays (SA Biosciences, Fredrick, MD, USA) as described previously.Citation14 Penl1 cells were seeded into a 96-well white plate and incubated overnight at 37°C. Transient transfection was conducted by adding plasmid construct of transcription factor-responsive reporter of each signaling pathway and controls to cells and incubated overnight in a 37°C incubator. Then, cells were further incubated for 48 hours. Each transfection condition was carried in triplicates. After 48 hours, the changes in expression of each pathway in cells with or without CEACAM19 depletion were determined by measuring the generated firefly and Renilla luminescent signals using the Dual-Glo Luciferase Assay system (Promega, Madison, WI, USA) on the Glomax machine (Promega). The relative luciferase units were determined by dividing the firefly to Renilla luciferase activity ratio. The luciferase activity of Scr shRNA control was regarded as 100%.
ELISA for MMP2 and MMP9
Cell supernatant was centrifuged at 12,000 g for 15 minutes at 4°C. MMP2 ELISA kits (Sigma) and MMP9 ELISA kits (Sigma) were used to measure the levels of secreted MMP2 and MMP9 by Penl1 cells, respectively, according to the manufacturer’s instructions.Citation24
Statistical analysis
The statistical software package SPSS 16.0 was used in this study. Descriptive statistics were used to summarize clinicopathological features. Survival analysis was performed using the Kaplan–Meier method to determine overall CSS. The log-rank test was used to compare survival curves. Multivariable Cox regression analysis was conducted to identify the prognostic factors that influence CSS. Single comparisons were performed using Student’s t-test. Two-sided P-values <0.05 were considered significant.
Results
CEACAM19 was differentially expressed in non-cancerous penile tissues and penile cancers
CEACAM19 expression in penile cancer tissues (n=64) was analyzed by immunohistochemical staining. In noncancerous penile tissues, CEACAM19 immunostaining was low or absent, and confined mostly in the basal cells of the epithelium (). CEACAM19 exhibited differential immunostaining in the penile cancer samples (Case 1–3, CEACAM19 over-expression; Case 4–6, CEACAM19 low expression). Overall, 30 (46.88%) cases showed CEACAM19 over-expression (staining positivity ≥30%). In most cases, CEACAM19 exhibited cytoplasmic and nuclear staining in penile cancer tissues (). The clinicopathological features of the study cohort are summarized in . As shown in , over-expression of CEACAM19 was significantly related to distant metastasis (P=0.021) and lymph node metastasis (P=0.000), but not to age (P=0.205), differentiation (P=0.676), histological subtype (P=0.420), T stage (P=0.380), or HPV status (P=0.384). We also conducted further analysis on the relationship between nodal status and other clinicopathological parameters. As shown in , clinicopathological parameters such as differentiation (P=0.048) and metastasis stage (P=0.001) were also significantly associated with nodal status. Kaplan–Meier survival analysis showed that over-expression of CEACAM19 was associated with unfavorable CSS (P=0.001, ). Consistent with previous studies, the status of lymph node metastasis could also predict the prognosis in our study cohort (P<0.001, ). We also conducted multivariable Cox regression analysis and the results were summarized in . Our results showed that HPV status (P=0.018), nodal status (P=0.030), histological subtype (P=0.015), and metastasis stage (P=0.003) could serve as independent predictor for CSS. However, CEACAM19 over-expression could not be an independent predictor of CSS (HR 2.29, 95% CI 0.709–7.400, P=0.166).
Figure 1 Notes: (A) Expression of CEACAM19 in non-cancer penile tissues and penile cancer. Bars: 100 µm. Upper panel, low expression of CEACAM19 in non-cancer penile tissues (1–3); middle panel, penile cancer cases with over-expression of CEACAM19 (Case 1–3); lower panel, penile cancer cases with low expression of CEACAM19 (Case 4–6). (B) Over-expression of CEACAM19 in penile cancer was associated with unfavorable CSS. (C) Nodal status predicted unfavorable CSS in penile cancer. The log-rank test was used to compare survival curves.
Abbreviation: CSS, cancer-specific survival.
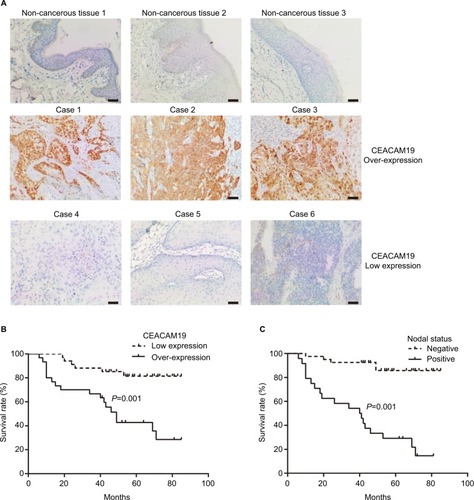
Table 1 Demographic and clinicopathological characteristic of study population associated with CEACAM19 expression
Table 2 Demographic and clinicopathological characteristic of study cohort associated with nodal status
Table 3 Cox proportional hazard model for prognosis associated with clinicopathological parameters and CEACAM19 expression
Knockdown of CEACAM19 expression suppresses cell growth and clonogenesis in Penl1 cells
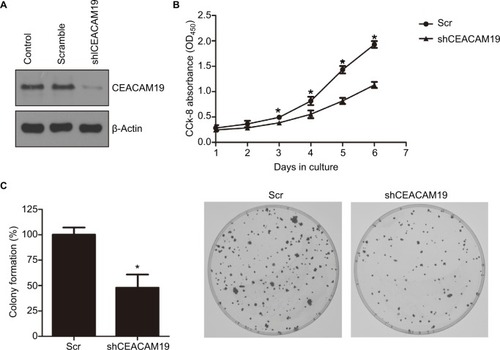
We sought to further investigate the oncogenic function of CEACAM19 using Penl1 as cell model of penile cancer. Penl1 cells were transduced with non-targeting Scr or specific shRNA targeting CEACAM19 (shCEACAM19) As shown in , CEACAM19 expression was significantly reduced in Penl1 cells transduced with shCEACAM19 lentivirus, whereas it was not significantly affected by Scr shRNA. We next examined the effect of CEACAM19 expression on the cell growth of Penl1 cells by CCK-8 assay, and the results showed that shCEACAM19 transduced-Penl1 cells grew slower than those transduced with Scr shRNA (P<0.05; ). Furthermore, colony formation of Penl1 cells in shCEACAM19 group decreased greatly, as compared with Scr group, (P<0.05; ).
Figure 2 Knockdown of CEACAM19 expression suppresses cell growth and clonogenesis in Penl1 cells.
Notes: (A) CEACAM19 expression was significantly reduced in Penl1 cells transduced with shCEACAM19 lentivirus. β-Actin served as a loading control. (B) Knockdown of CEACAM19 expression attenuated cell growth of Penl cells. The CCK-8 absorbance was measured at 450 nm (OD450). *P<0.05, Scr vs shCEACAM19. (C) Depletion of CEACAM19 expression reduced clonogenesis of Penl1 cells. The colony formed in Scr control was regarded as 100%. *P<0.05, Scr vs shCEACAM19. All experiments were performed three times, and data are presented as mean ± SD. values. *P<0.05, Scr vs shCEACAM19.
Abbreviations: CCK-8, Cell Counting Kit-8; Scr, scramble; shCEACAM19, shRNA targeting CEACAM19.
Depletion of CEACAM19 expression attenuates cell migration and invasion in Penl1 cells
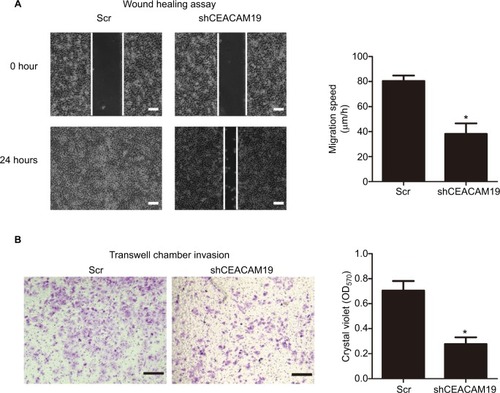
The metastatic process involves cell scattering, motility, ECM degradation, migration, and invasion through the basement membranes.Citation19–Citation21 Since over-expression of CEACAM19 was associated with nodal/distant metastasis, we proposed that CEACAM19 might regulate the migration/invasion phenotypes in penile cancer. To examine the effect of CEACAM19 on cancer cell migration in vitro, we performed wound healing assay. We observed a significantly slower wound-healing rate in the cells expressing shCEACAM19 shRNA compared to the Scr control (P<0.05; ). Transwell invasion assay was also performed in order to examine the effects of CEACAM19 on cell invasion. As shown in , knockdown of CEACAM19 expression attenuated the invasion of Penl1 cells, as compared with Scr control (P<0.05).
Figure 3 Depletion of CEACAM19 expression inhibits cell migration and invasion in Penl1 cells.
Notes: (A) Wound healing assay. The migration of the cells to the wound was measured at 0 and 24 hours after scratch. The representative fields were photographed; the relative healing rates were quantified with measurements of the gap sizes after the culture. Three different areas in each assay were chosen to measure the distances of migrating cells to the origin of the wound. Bars: 100 µm. *P<0.05, Scr vs shCEACAM19. (B) Transwell invasion assays with Penl1 cells were performed in Scr control and shCEACAM19 group. Crystal violet assay (OD570) was conducted to evaluate cell migration and invasion capability. Bars: 100 µm. All experiments were performed three times, and data are presented as mean ± SD. values. *P<0.05, Scr vs shCEACAM19.
Abbreviations: Scr, scramble; shCEACAM19, shRNA targeting CEACAM19.
CEACAM19 regulates oncogenic and metastasis-related signaling pathways in Penl1 cells
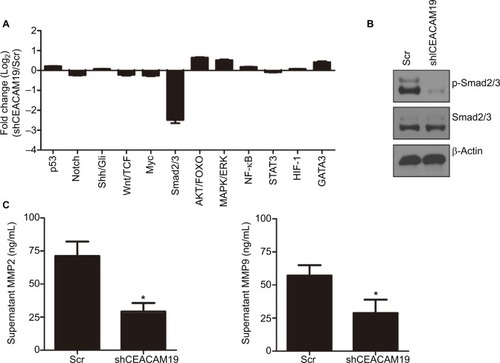
We conducted luciferase reporter assay on cancer-related signaling pathways in penl1 cells. As shown in , knockdown of CEACAM19 led to a dramatic decline of luciferase reporter activity in Smad2/3 pathways (|Log2 (fold change)|>2), as compared with Scr control (P<0.05). The reporter activities of other pathways such as p53, Notch, Shh/Gli displayed mild change in CEACAM19-depleted Penl1 cells (|Log2 (fold change)|<1). Western blotting analysis confirmed the reduction of p-Smad2/3 levels in CEACAM19-depleted Penl1 cells (). Furthermore, ELISA revealed that depletion of CEACAM19 reduced secretion of two invasion/metastasis-related molecules MMP2 and MMP9 in Penl1 cells (P<0.05; ).
Figure 4 Notes: (A) Multiple pathway reporter analysis identified potential cancer-related pathways regulated by CEACAM19 in Penl1 cells. (B) Knockdown of CEACAM19 expression attenuated p-Smad2/3 expression in Penl1 cells. β-Actin served as a loading control. (C) Knockdown of CEACAM19 expression reduced MMP2/9 secretion in Penl1 cells. All experiments were performed three times, and data are presented as mean ± SD values. *P<0.05, Scr vs sh CEACAM19.
Inhibition on Smad2/3 activation suppressed cell migration and invasion and reduced MMP2/9 secretion
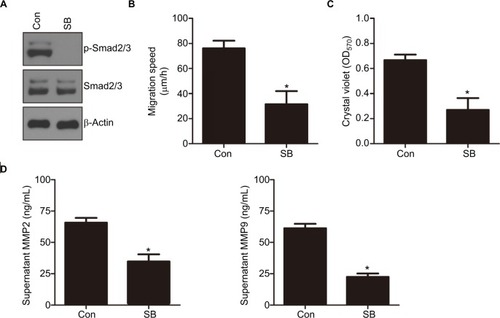
We showed that knockdown of CEACAM19 led to a decline of Smad2/3 activity in Penl1 cells. Therefore, Smad2/3 signaling might be important in mediating CEACAM19 effect on cell migration and invasion. As shown in , p-Smad2/3 level was potently reduced by TGF-β/Smad2/3 pathway inhibitor SB431542 (Cell Signaling). Furthermore, SB431542 suppressed cell migration and invasion in Penl1 cells (P<0.05; and ). ELISA revealed that incubation with SB431542 reduced secretion of MMP2 and MMP9 in Penl1 cells (P<0.05; ).
Figure 5 Notes: (A) The effect of SB431542 (SB) on p-Smad2/3 levels in Penl1 cells. Penl1 cells were treated with 10 µM SB for 24 hours. β-Actin served as a loading control. (B) The effect of SB431542 on wound healing of Penl1 cells. Three different areas in each assay were chosen to measure the distances of migrating cells to the origin of the wound. *P<0.05, Control vs SB. (C) The effect of SB431542 on cell invasion of Penl1 cells. Transwell invasion assay was conducted to evaluate cell migration and invasion capability. *P<0.05, Control vs SB. (D) Inhibition on TGF-β/Smad2/3 activity by SB431542 reduced MMP2/9 secretion in Penl1 cells. All experiments were performed three times, and data are presented as mean ± SD. values. *P<0.05, Control vs SB.
Discussion
The primary treatment for penile cancer is surgery.Citation25 Although surgery alone can cure most of patients with local disease, the clinical outcome for patients with advanced penile cancer still remains poor.Citation26 Regional lymph node metastasis is the most important prognostic factor for predicting cancer-related survival.Citation26 Other clinicopathological factors such as HPV status and histological subtype could also serve as predictor for patient survival in penile cancer.Citation26–Citation28 Prognostic biomarkers for the development of lymph node metastasis are being intensively investigated. Squamous cell carcinoma antigen, proliferating cell nuclear antigen, cyclin D1, as well as other markers have been studied with limited clinical utility in penile cancer.Citation6 However, none of them can predict lymph node status or exhibit prognostic value.Citation6 It is urgent to identify reliable biomarkers for the early diagnosis of nodal metastasis of penile cancer.
Several biomarkers have been under clinical investigation for their potential of predicting lymph node metastasis. Lopes et al showed that p53 expression could serve as a new prognostic factor for lymph node metastasis in penile carcinoma.Citation29 Ferrandiz-Pulido et al recently unveiled the mammalian target of rapamycin pathway in the development of lymph node metastasis.Citation30 Poetsch et al indicated that loss of tumor suppressor gene p16INK4A is associated with aggressive behavior of penile carcinomas; elevated C-reactive protein values predict nodal metastasis in patients with penile cancer.Citation31,Citation32 In this study, we evaluated CEACAM19 expression in penile cancer and analyzed its association with clinicopathological features. We showed that CEACAM19 is differentially expressed in non-cancerous tissues and penile cancer; CEACAM19 over-expression was significantly associated with nodal and distant metastasis, and could serve as a prognostic factor to predict unfavorable CSS in penile cancer. However, due to the limits of small cohort (n=64) enrolled in this study, larger cohort would be warranted in order to further elucidate the usefulness of CEACAM19 as a biomarker for the prediction of prognosis or nodal status in penile cancer.
Tumor progression is mechanistically driven by alterations in the regulatory mechanisms of proliferation and migration/invasion. However, the molecular alterations involved in penile cancer progression still await investigation, although significant advances have been made in discovering the molecular pathways driving tumor progression. It has previously shown that CEACAM19 was over-expressed in breast cancer tissues, and was proposed as a key factor of tumor progression.Citation11,Citation12 Zhao et al found that CEACAM19 was over-expressed in gastric cancer.Citation33 However, the function of CEACAM19 in the malignant progression of penile cancer still remains unclear.
In a series of experiments in penile cancer cell line model, we showed that depletion of CEACAM19 attenuated the cell proliferation and clonogenesis in Penl1 cells, highlighting the important role of CEACAM19 in the regulation of tumorigenesis in penile cancer. Our study also revealed the critical role of CEACAM19 in maintaining the metastatic phenotype of penile cancer, as knockdown of CEACAM19 impaired the migration and invasiveness of penl1 cells. Furthermore, depletion of CEACAM19 markedly attenuated p-Smad2/3 in Penl1 cells, suggesting CEACAM19 might exert its oncogenic function via regulating the activation of Smad2/3 in penile cancer cells. As activation of TGF-β/Smad2/3 pathway has already been demonstrated to be involved in the malignant progression and dissemination of various cancer types, it would be reasonable to propose that the effects of CEACAM19 might partly result from its function in regulating the Smad2/3 activation, thus promoting the metastasis of penile cancer.Citation34 Soares et al showed that penile cancers with over-expression of two metastasis-related molecules MMP2 and MMP9 were deeply invasive.Citation35 Similarly, we observed that the secretion of MMP2 and MMP9 was markedly down-regulated in Penl1 cells with CEACAM19 knockdown or treated with TGF-β/Smad2/3 inhibitor SB431542, suggesting CEACAM19 might regulate the metastatic phenotype of penile cancer via modulating Smad2/3 and its downstream MMP2/9 expression. Although high expression of p53 was previously shown to be significantly associated with metastasis and poor survival in penile cancer,Citation36,Citation37 we did not observe considerable reduction of p53 activities in CEACAM19-depleted cells, suggesting the mechanism of CEACAM19 might be independent of p53 function.
Conclusion
In this study, we showed that over-expression of CEACAM19 was significantly associated with nodal and distant metastasis, and predicted unfavorable CSS in penile cancer. Based on in vitro cell line model study, we showed that depletion of CEACAM19 expression suppressed cell proliferation, reduced colony formation, and attenuated cell migration and invasion in Penl1 cells. Furthermore, we showed that the effects of CEACAM19 might result from its function in regulating the Smad2/3 activation, thus promoting cell migration and invasion, and stimulating MMP2/9 secretion in penile cancer cells. Cumulatively, our findings would highlight the importance of CEACAM19 in the regulation of cell proliferation and metastasis of penile cancer, and suggest that CEACAM19 expression might serve as a valuable prognostic biomarker and therapeutic target for patients with penile cancers. Future prospective study with a greater number of patients would be required to validate the clinical usefulness of CEACAM19 in penile cancer.
Acknowledgments
This work was supported by National Nature Science Foundation of China (81672510).
Disclosure
The authors report no conflicts of interests in this work.
References
- PanerGPStadlerWMHanselDEMontironiRLinDWAminMBUpdates in the eighth edition of the tumor-node-metastasis staging classification for urologic cancersEur Urol201873456056929325693
- DouglawiAMastersonTAUpdates on the epidemiology and risk factors for penile cancerTransl Androl Urol20176578579029184774
- OttenhofSRLeoneARHorenblasSSpiessPEVegtEAdvancements in staging and imaging for penile cancerCurr Opin Urol201727661262028937510
- BarskiDGeorgasEGerullisHEckeTMetastatic penile carcinoma -an update on the current diagnosis and treatment optionsCent European J Urol2014672126132
- FicarraVNovaraGBoscolo-BertoRArtibaniWKattanMWHow accurate are present risk group assignment tools in penile cancer?World J Urol200927215516018560836
- Zargar-ShoshtariKSharmaPSpiessPEInsight into novel biomarkers in penile cancer: redefining the present and future treatment paradigm?Urol Oncol2018361043343929103967
- StankiewiczENgMCuzickJThe prognostic value of Ki-67 expression in penile squamous cell carcinomaJ Clin Pathol201265653453722447920
- MannweilerSSygullaSWinterERegauerSTwo major pathways of penile carcinogenesis: HPV-induced penile cancers overexpress p16ink4a, HPV-negative cancers associated with dermatoses express p53, but lack p16ink4a overexpressionJ Am Acad Dermatol2013691738123474228
- AryaMThrasivoulouCHenriqueRTargets of Wnt/β-catenin transcription in penile carcinomaPLoS One2015104e012439525901368
- ScorilasAChiangPMKatsarosDYousefGMDiamandisEPMolecular characterization of a new gene, CEAL1, encoding for a carcinoembryonic antigen-like protein with a highly conserved domain of eukaryotic translation initiation factorsGene2003310798912801635
- MichaelidouKTzovarasAMissitzisIArdavanisAScorilasAThe expression of the CEACAM19 gene, a novel member of the CEA family, is associated with breast cancer progressionInt J Oncol20134251770177723525470
- EstiarMAEsmaeiliRZareAAHigh expression of CEACAM19, a new member of carcinoembryonic antigen gene family, in patients with breast cancerClin Exp Med201717454755327909883
- ChenJYaoKLiZEstablishment and characterization of a penile cancer cell line, penl1, with a deleterious TP53 mutation as a paradigm of HPV-negative penile carcinogenesisOncotarget2016732516875169827351128
- JunFHongJLiuQEpithelial membrane protein 3 regulates TGF-β signaling activation in CD44-high glioblastomaOncotarget201789143431435827527869
- WolffACHammondMESchwartzJNAmerican Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancerJ Clin Oncol200725111814517159189
- FarajSFChauxAGonzalez-RoibonNImmunohistochemical expression of ARID1A in penile squamous cell carcinomas: a tissue microarray study of 112 casesHum Pathol201546576176625776029
- YangYHuangJQZhangXShenLFMiR-129-2 functions as a tumor suppressor in glioma cells by targeting HMGB1 and is down-regulated by DNA methylationMol Cell Biochem20154041–222923925772485
- LiPTianWMaXAlpha-mangostin inhibits intracellular fatty acid synthase and induces apoptosis in breast cancer cellsMol Cancer20141313824894151
- YangCXuYChengFmiR-1301 inhibits hepatocellular carcinoma cell migration, invasion, and angiogenesis by decreasing Wnt/β-catenin signaling through targeting BCL9Cell Death Dis201788e299928817119
- HeJXuQWangMOral administration of apigenin inhibits metastasis through AKT/P70S6K1/MMP-9 pathway in orthotopic ovarian tumor modelInt J Mol Sci20121367271728222837693
- ShanNShenLWangJHeDDuanCMiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19Biochem Biophys Res Commun2015456138539125475731
- FangFChangRMYuLMicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinomaJ Hepatol201563487488525998163
- ZhangJFuJPanYZhangXShenLSilencing of miR-1247 by DNA methylation promoted non-small-cell lung cancer cell invasion and migration by effects of STMN1Onco Targets Ther201697297730727942223
- HuJNiSCaoYThe angiogenic effect of microRNA-21 targeting TIMP3 through the regulation of MMP2 and MMP9PLoS One2016112e014953726872030
- HuXHuangJWenSFuJChenMComparison of efficacy between brachytherapy and penectomy in patients with penile cancer: a meta-analysisOncotarget201785910046910047729245993
- WangJYGaoMZYuDXHistological subtype is a significant predictor for inguinal lymph node metastasis in patients with penile squamous cell carcinomaAsian J Androl201820326526929286007
- BezerraSMChauxABallMWHuman papillomavirus infection and immunohistochemical p16(INK4a) expression as predictors of outcome in penile squamous cell carcinomasHum Pathol201546453254025661481
- CubillaALThe role of pathologic prognostic factors in squamous cell carcinoma of the penisWorld J Urol200927216917718766352
- LopesABezerraALPintoCASerranoSVde MelloCAVillaLLp53 as a new prognostic factor for lymph node metastasis in penile carcinoma: analysis of 82 patients treated with amputation and bilateral lymphadenectomyJ Urol20021681818612050497
- Ferrandiz-PulidoCMasferrerETollAmTOR signaling pathway in penile squamous cell carcinoma: pmTOR and peIF4E over expression correlate with aggressive tumor behaviorJ Urol201319062288229523764082
- PoetschMHemmerichMKakiesCAlterations in the tumor suppressor gene p16(INK4A) are associated with aggressive behavior of penile carcinomasVirchows Arch2011458222122921085986
- Al GhazalASteffensSSteinestelJElevated C-reactive protein values predict nodal metastasis in patients with penile cancerBMC Urol2013135324148787
- ZhaoHXuJWangYKnockdown of CEACAM19 suppresses human gastric cancer through inhibition of PI3K/Akt and NF-κBSurg Oncol201827349550230217308
- BellomoCCajaLMoustakasATransforming growth factor β as regulator of cancer stemness and metastasisBr J Cancer2016115776176927537386
- SoaresFAda CunhaIWGuimarãesGCNonogakiSCamposRSLopesAThe expression of metaloproteinases-2 and -9 is different according to the patterns of growth and invasion in squamous cell carcinoma of the penisVirchows Arch2006449663764617072641
- ZhuYZhouXYYaoXDDaiBYeDWThe prognostic significance of p53, Ki-67, epithelial cadherin and matrix metalloproteinase-9 in penile squamous cell carcinoma treated with surgeryBJU Int2007100120420817433031
- GuniaSKakiesCErbersdoblerAHakenbergOWKochSMayMExpression of p53, p21 and cyclin D1 in penile cancer: p53 predicts poor prognosisJ Clin Pathol201265323223622135025
