Abstract
Background
Growing evidence has indicated that the long noncoding RNA H19 (lncRNA H19), frequently deregulated in almost all tumor types tested, acted as a pivotal contributor to both cancer initiation and progression. However, the role of lncRNA H19 in human papillary thyroid carcinoma (PTC) remains controversial. The aim of the study was to investigate the expression and potential function of lncRNA H19 in human PTC.
Patients and methods
The lncRNA H19 level was determined by quantitative real-time (RT)-PCR analyses in 58 PTC tissue samples and their paired paracancerous tissue samples. RNA interference, RT-PCR analysis, and Western blot assay were used to determine the impact of lncRNA H19 on epithelial-mesenchymal transition (EMT) markers in human PTC cells. The migratory and invasive capacities of PTC cells were determined by wound-healing and transwell migration and invasion assays.
Results
lncRNA H19 expression was 2.417-fold higher in PTC tissues than their paired paracancerous tissue (95% CI: 1.898–2.935, P<0.0001). Higher level of lncRNA H19 was correlated to elevated expression of Vimentin, ZEB2, Twist, and Snail2. Inhibition of lncRNA H19 resulted in upregulation of E-cadherin and downregulation of Vimentin both at mRNA and protein levels. Conversely, enforced expression of the exogenous lncRNA H19 led to E-cadherin mRNA and protein downregulation and relative upregulation of Vimentin. Moreover, wound-healing and transwell migration and invasion assays showed that lncRNA H19 could promote the migratory and invasive abilities of PTC cells.
Conclusion
The level of lncRNA H19 was significantly higher in PTC tissues than paired paracancerous tissue or normal tissues. Overexpression of lncRNA H19 was correlated with higher tumor burden of PTC. It also contributes to EMT process, as well as promotes migration and invasion of PTC cells.
Introduction
Growing concern over the sharp increase in the prevalence of thyroid cancer in past decades, particularly papillary thyroid carcinoma (PTC), has prompted investigators to explore the molecular underpinnings of the disease, leading to the identification of BRAFV600E and RAS mutations, as valid therapeutic targets. Citation1–Citation3 Recently, the Encyclopedia of DNA Elements project, which was designed to discover and characterize novel RNAs transcripts, has revealed that a large amount of primary or processed protein-coding and noncoding transcripts, including thousands of long noncoding RNAs (lncRNAs), are predominantly implicated in the human genome. Citation4,Citation5 Notably, increasing studies have demonstrated that deregulated expression of lncRNAs was significantly involved in the pathogenesis of multiple types of cancer. Citation6,Citation7
The lncRNAs are a new class of nonprotein transcripts with over 200 bases in length. Citation8 Emerging evidence has indicated that the lncRNA H19, frequently deregulated in almost all tumor types tested, acted as a critical contributor to both cancer initiation and progression. Citation9 Li et al reported that overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Citation10 Guan et al reported that overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Citation11 A study by Shi et al demonstrated that lncRNA H19 predicts poor prognosis in patients with melanoma and regulates cell growth, invasion, migration, and epithelial-mesenchymal transition (EMT) in melanoma cells. Citation12 Moreover, preclinical and clinical studies have highlighted the therapeutic potential against lncRNA H19. For instance, Zhao et al showed that downregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-κB pathway. Citation13
Despite enormous breakthroughs in the understanding of the role of lncRNA H19 in multiple types of tumor progression, the exact role and function of lncRNA H19 in human PTC remain under-investigated. A major mechanism through which BRAF mutation might contribute to primary PTC progression is through induction of EMT. Citation14,Citation15 Specifically, the Snail/E-cadherin axis has been demonstrated as a pathway dysregulated by BRAF, leading to EMT. Citation16 The possibility that lncRNA H19 can regulate EMT program is an attractive option to explore. In this study, we identified that the lncRNA H19 was significantly increased in PTC tissues and associated with classic EMT markers in PTCs, suggesting that lncRNA H19 may be required for human PTC tumorigenesis and development.
Patients and methods
Patients and clinical tissue samples
Written informed consents were received from all patients before enrollment, and this study was approved by the Ethics Committee of Shantou University Medical Hospital. Fifty-eight PTC tissue samples and their paired paracancerous tissue samples were obtained from patients (age range: 27–72 years; 16 males and 42 females), who were first diagnosed between June 2016 to December 2017 at the First Affiliated Hospital of Shantou University Medical Hospital for quantitative real-time (qRT)-PCR analyses and paraffin-embedded pathological investigation. All medical histories of the patients were well-documented according to the eighth edition of the American Joint Committee on Cancer TNM system. Citation17
Cell lines and cell culture
The human PTC cell line B-CPAP was purchased from the Chinese Academy of Sciences (Beijing, China). This cell line was established from the tumor tissue of a woman aged 72 years with metastatic PTC in 1992. Citation18 Cells were cultured in RPMI-1640 medium, containing 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin (all from Thermo Fisher Scientific, Waltham, MA, USA) in a 5% CO2 incubator at 37°C.
RNA purification and RT-PCR analysis
All obtained tissues during surgery were immediately stored in liquid nitrogen prior to use. Total RNA (1 µg) was isolated from tissues or cells using TRIzol (Thermo Fisher Scientific) following the manufacturer’s instructions and stored at −80°C. Reverse transcription was performed using a Prime-Script™ RT reagent kit (Takara Bio Inc., Shiga Prefecture, Japan) according to the manufacturer’s instructions. To detect mRNA expression, qRT-PCR was performed with an SYBR Select Master Mix (Thermo Fisher Scientific) and a CFX96 RT-PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA). To normalize the amount of mRNA in each sample, GAPDH was used. Primer sequences used in RT-PCR are listed in .
Small interfering RNAs (siRNAs) and transfection
The siRNAs were purchased from Suzhou GenePharma Co., Ltd. (Suzhou, China). The siRNAs are shown in . Cells were transfected using Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA). A total of 1×106 B-CPAP cells were inoculated into each well of a 6-well culture plate. The next day, 75 pmol of siRNA was combined with 3.75 µL of Lipofectamine 3000 (Life Technologies) by strictly following the manufacturer’s instructions.
Western blot assay
Cells were lysed in RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) with protease inhibitors. The extracted protein sample was measured using the BCA protein assay and was stored at −80°C until required for use. The 30 µg protein sample was separated by 8% SDS-PAGE and transferred onto a PVDF membrane (EMD Millipore, Billerica, MA, USA). After transferring, the membrane was blocked in 5% nonfat milk and then incubated with primary antibodies at 4°C overnight. The antibodies are shown in . The next day, the membrane was incubated with secondary antibodies and subjected to electrochemiluminescence detection (Applygen Technologies, Inc., Beijing, China).
Wound healing assay
Thirty minutes before the application of injury lines, B-CPAP cells were treated with Mitomycin C (25 mg/mL) (Beyotime Institute of Biotechnology, Jiangsu, China). Injury line was made with a 2-mm wide tip separated on cells plated in culture dishes at 90% confluency. After washing with PBS, cells could migrate in complete medium and photographs were taken (×40) after 48 hours. An average of five random widths of each injury line was measured for quantitation.
Transwell invasion assays
Cell culture inserts (8 M pore size; BD, Franklin Lakes, NJ, USA) and Matrigel invasion chambers (BD) were used to perform migration and invasion assays, respectively. Transfected cells were serum-starved for 24 hours and then 3×104 B-CPAP cells in serum-free medium were seeded into the upper chamber. Complete medium was added to the bottom chamber. Cells were stained with 0.1% crystal violet for migration assays after 36 and 48 hours for invasion assays. Each assay was performed in triplicate. The number of cells from five fields in each well were counted by two investigators (HYL and DZ).
Clinical databases
The ONCOMINE datasets (http://www.oncomine.org/) were used to analyze the expression of lncRNA H19 in various cancers against normal tissue as well as the correlated expression genes in thyroid cancer. Citation19 The correlations between the expression levels of lncRNA H19 and Vimentin, ZEB2, Twist, or Snail2 in thyroid cancers were determined through analysis in the cBioPortal database (http://www.cbioportal.org/index.do). Citation20 We analyzed thyroid carcinoma (TCGA, Provisional) and PTC (TCGA, Cell 2014) datasets. The expression levels of lncRNA H19 in a series of cancers were analyzed by CCLE database (https://portals.broadinstitute.org/ccle/home), which is an online encyclopedia of a compilation of gene expression, chromosomal copy number, and massively parallel sequencing data from 947 human cancer cell lines. Citation21
Statistical analysis
Statistical analysis was performed using the SPSS version 18.0. Data from three independent experiments were presented as the mean ± SD. Statistical analysis was performed using Mann–Whitney U test and Wilcoxon signed-rank test. The clinicopathological characteristics were analyzed using Pear-son’s chi-squared test. Difference between multiple groups was analyzed by two-way ANOVA. For clinical data from ONCOMINE and cBioPortal datasets, Student’s t-test was used, and two times of fold change with a P-value of <0.01 was defined as clinically significant, Correlation coefficients between RNA levels were obtained through Spearman’s rank correlation analysis as was described earlier. Citation22 A value of P<0.05 was considered as a statistically significant.
Ethical statement
The study was approved by the ethics committee of the First Hospital of Shantou University Medical College. Patient consent was written informed consent, in compliance with the Declaration of Helsinki.
Results
The expression level of lncRNA H19 was significantly increased in PTC tissues
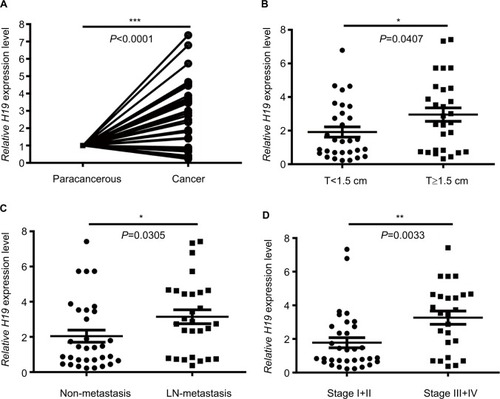
lncRNA H19 has been identified as an oncogenic gene in multiple cancer types and its elevated expression was tightly linked to tumorigenesis and cancer progression. ONCO-MINE analysis revealed that lncRNA H19 expression was significantly higher in cancer than normal samples across a wide variety of datasets in different cancer types. A total of 220 different types of studies about lncRNA H19 were collected in the ONCOMINE database () in which 37 studies showed the increase of lncRNA H19 expression and 22 studies showed a decrease. In addition, CCLE analysis demonstrated that the expression level of lncRNA H19 in thyroid cancer cells ranked the eleventh highest in a variety of cancer cell lines tested (). Thus, we performed the qRT-PCR assay to detected PTC tissue samples and corresponding adjacent nontumorous tissues samples from 58 patients who were first diagnosed in our hospital. Our data showed that lncRNA H19 expression was 2.417-fold higher (95% CI: 1.898–2.935) in PTC tissues than their paired paracancerous tissues (, P<0.0001). These results implied that lncRNA H19 may be distinctively expressed higher in thyroid cancer, suggesting it might play unique roles in the development of thyroid cancer.
Figure 1 Analysis of lncRNA H19 expression in PTC patients according to qRT-PCR.
Notes: (A) Relative expression of H19 in cancer group and paracancerous group. (B) Relative expression of H19 in T<1.5 cm group and T≥1.5 cm group. (C) Relative expression of H19 in non-metastasis group and lymph node metastasis group. (D) Relative expression of H19 in stage I+II group and stage III+IV group (*P<0.05, **P<0.01, and ***P<0.001).
Abbreviations: lncRNA H19, long noncoding RNA H19; PTC, papillary thyroid carcinoma; qRT-PCR, quantitative real-time PCR.
Elevated lncRNA H19 expression correlated with tumor burden of PTC
To further investigate the correlation between lncRNA H19 expression and clinicopathological features, qRT-PCR analyses of lncRNA H19 were performed using samples from 58 PTC patients. When the 58 tumor tissues were stratified on clinical progression, we found that higher lncRNA H19 expression is remarkably associated with increased tumor size (, P=0.0407). In addition, lncRNA H19 expression levels were much higher in lymph node metastasis group than those in patients without lymph node metastasis (, P=0.0305). Moreover, high lncRNA H19 expression was correlated with PTC patients of stage III/IV than those in patients with stage I/II (, P=0.0033). Consistently, when 58 PTC patients were divided into two group, lncRNA H19High (>median), and lncRNA H19Low (<median), our data also revealed that more patients in increased tumor size group (P=0.036), lymph node metastasis group (P=0.002), and higher stage group (P=0.004) correlated with lncRNA H19High group (). However, the level of lncRNA H19 expression was not associated with age, gender, extrathyroidal extension status, pathological subtype, and whether combined with nodular goiter between these two groups (). It was, therefore, postulated that the high lncRNA H19 expression level was correlated with more malignant phenotypes with increased proliferation and potential of metastasis in PTC.
Table 1 Clinicopathological characteristics of 58 PTC patients according to H19 expression
Higher expression of lncRNA H19 correlated with elevated expression of mesenchymal phenotypes markers Vimentin, ZEB2, Twist, and Snail2
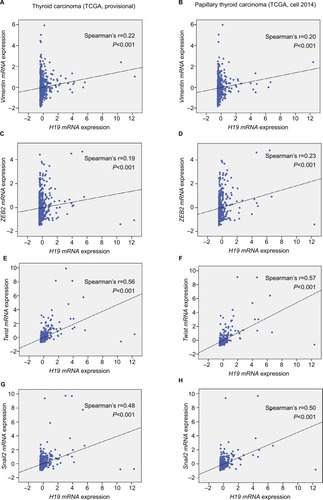
To explore the inherent properties of lncRNA H19 and its potential mechanism in PTC, we used the ONCOMINE and cBioPortal databases to analyze the correlation of lncRNA H19 in connection with other featured biomarkers in PTC. Interestingly, in ONCOMINE co-expression analysis, it was found that lncRNA H19 expression was significantly correlated with the expression of ZEB2, a typical mesenchymal phenotypes marker (, r=0.700). Moreover, gene correlation targeted analysis from TCGA database showed that in thyroid carcinoma, higher expression of lncRNA H19 correlated with elevated mesenchymal biomarker Vimentin (, r=0.22, P<0.001), ZEB2 (, r=0.19, P<0.001), Twist (, r=0.56, P<0.001), and Snail2 (, r=0.48, P<0.001). Meanwhile, correlation analysis in PTC showed that elevated lncRNA H19 correlated with increased expression of Vimentin (, r=0.20, P<0.001), ZEB2 (, r=0.23, P<0.001), Twist (, r=0.57, P<0.001), and Snail2 (, r=0.50, P<0.001). These results showed that the expression of lncRNA H19 in PTC positively associated with mesenchymal phenotypes marker, which imply that lncRNA H19 might play a distinct role in the processing of EMT and lead to more aggressive clinicopathological features of PTC.
Figure 2 H19 expression was significantly correlated with ZEB2 expression in thyroid carcinoma (shown in red frame, from ONCOMINE correlation analysis).
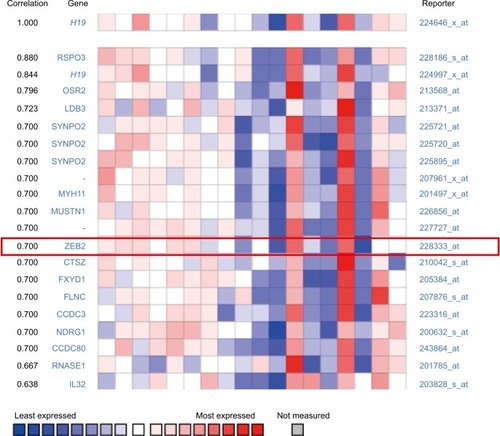
Figure 3 Elevated H19 expression correlated with higher expression of Vimentin, ZEB2, Twist, and Snail2 in thyroid carcinoma.
Notes: In the TCGA (the Cancer Genome Atlas) database, the Pearson’s chi-squared test was performed to analyze the correlation of H19 and some typical mesenchymal markers. (A and B) Gene correlation targeted analysis between H19 and Vimentin in samples of (A) thyroid carcinoma or (B) PTC. (C and D) Gene correlation targeted analysis between H19 and ZEB2 in samples of (C) thyroid carcinoma or (D) PTC. (E and F) Gene correlation targeted analysis between H19 and Twist in samples of (E) thyroid carcinoma or (F) PTC. (G and H) Gene correlation targeted analysis between H19 and Snail2 in samples of (G) thyroid carcinoma or (H) PTC.
Abbreviation: PTC, human papillary thyroid carcinoma.
lncRNA H19 regulates mRNA and protein expression of EMT markers E-Cadherin and Vimentin
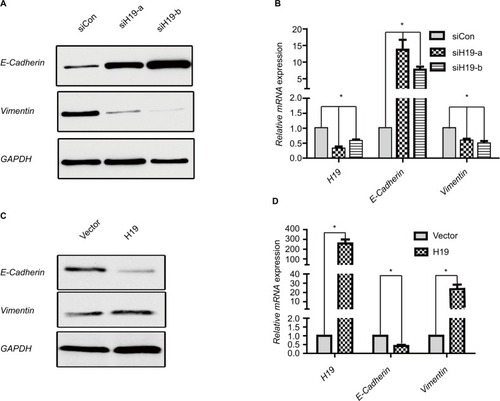
Since the above data from the public database and clinical sample have shown that H19 was correlated with EMT in PTC, we verified whether lncRNA H19 was associated with typical EMT markers in PTC cell line B-CPAP. Suppression of lncRNA H19 by siRNA caused a significant upregulation of epithelial marker E-cadherin expression at both the mRNA and protein levels, while that of Vimentin, a mesenchymal marker, decreased as compared to controls ( and ). Two different siRNA sequences were used to silence lncRNA H19 separately to exclude the possibility of nonspecific target. It was found that siH19-a and siH19-b effectively suppressed lncRNA H19 and finally caused downregulated Vimentin, while upregulating E-cadherin both at mRNA and protein levels ( and ). We chose siH19-b with better suppression effect on lncRNA H19 for further experiments. Conversely, enforced expression of the exogenous lncRNA H19 led to E-cadherin mRNA and protein downregulation and relative upregulation of Vimentin ( and ). These results implied that lncRNA H19 was able to mediate the process of EMT in PTC cells.
Figure 4 H19 regulates expression of EMT marker E-cadherin and Vimentin in PTC cells.
Notes: (A and B) Expression of E-cadherin and Vimentin in B-CPAP cells at protein and mRNA levels analyzed by (A) Western blot and (B) RT-PCR, respectively, when silencing H19 by siH19-a and siH19-b. (C and D) Expression of E-cadherin and Vimentin in B-CPAP cells at protein and mRNA levels analyzed by (C) Western blot and (D) RT-PCR, respectively, when overexpressing H19. *P<0.05 as compared to control groups.
Abbreviations: EMT, epithelial-mesenchymal transition; PTC, papillary thyroid carcinoma; RT-PCR, real-time PCR.
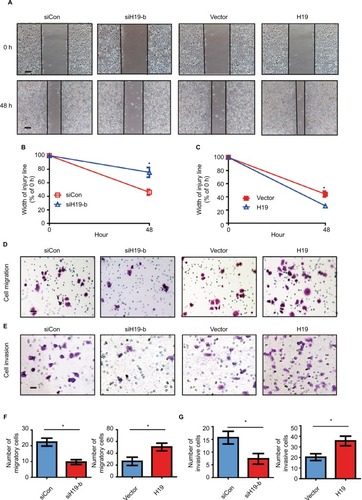
lncRNA H19 promotes migration and invasion abilities of PTC cells
To evaluate whether the expression of lncRNA H19 regulates cellular function via inducing EMT, we performed gain-of-function and loss-of-function studies on B-CPAP cells. We first assessed the effect of lncRNA H19 knockdown on cell motility via a wound healing assay. As displayed in , after culturing for 48 hours, the width of the scratch wound in siH19-b B-CPAP cells had reduced to 75% as compared to 0 hours, while the width of the scratch wound in control cells had reduced to 46% as compared to 0 hours (), indicating that loss of lncRNA H19 expression inhibits cell motility. In contrast, when enforcing expression of the exogenous lncRNA H19 in B-CPAP cells, the scratch wound width had healed to 26% as compared to 0 hours, dwarfing the 44% gap healed by control cells. This result suggests that lncRNA H19 overexpression was capable of promoting the motility of B-CPAP cells (). We further investigated whether lncRNA H19 influenced the migration and invasion abilities of B-CPAP cells via transwell migration and invasion assays. As shown in , the number of migrated cells transfected with siH19 was significantly lower (~52%) than the number of migrated cells transfected with si-control (). Similarly, invasion assays showed that knocking down lncRNA H19 via siRNAs resulted in fewer cells invading through the Matrigel-coated membrane ( and ). Moreover, the ectopic expression of lncRNA H19 in B-CPAP cells promoted cellular migration and invasion (). These results indicate that lncRNA H19 enhances the migration and invasion abilities of PTC cells.
Figure 5 H19 knockdown in B-CPAP cells inhibits EMT while overexpression promotes EMT.
Notes: (A–C) Wound healing assay showed the knockdown of H19 inhibited the cellular motility of B-CPAP cells, while overexpression of H19 enhanced such motility. (A) Representative pictures and (B and C) quantitative data of wound recovery after 48 hours cell culture. Each sample contained three wells. (D–G) Knockdown of H19 decreased the cellular migration and invasion of B-CPAP cells; while overexpression of H19 enhanced such motility. (D) Representative pictures of migration and (E) invasion, (F) quantitative data of migration, or (G) invasion assays. All experiments were performed at least thrice and data were statistically analyzed by two-way ANOVA. *P<0.05 vs control. Error bars indicate SD.
Abbreviation: EMT, epithelial-mesenchymal transition.
Discussion
Thyroid cancer is the most common endocrine malignancy across the world. Citation1 Its prevalence has been steadily increasing in the past few decades, primarily due to a remarkable increase in the incidence of PTC. Citation23 Multiple patterns of genetic alteration, including BRAF mutations, RAS mutations, and RET/PTC rearrangements, have been identified to be critically involved in the tumorigenesis and progression of PTCs. Citation24–Citation26 Targeting therapies toward these genetic alterations have led to a significant improvement in the therapeutic response and survival of patients with PTCs. Citation27 Except for a series of protein-coding genetic alteration, a variety of noncoding transcripts have also been found to play essential roles in the development of a number of solid tumors. Citation28–Citation30 However, up to now, the alteration in the noncoding transcript in PTCs remained under-investigated. It has been recognized in most studies that lncRNA H19 overexpresses and contributes to tumorigenesis and progression of multiple cancer types, including gastric cancer, glioma, and head and neck squamous cell carcinoma. Citation10,Citation11,Citation31 Nevertheless, it was found that lncRNA H19 was under-expressed in intratumoral hepatocellular carcinoma (HCC) tissues, as compared to peritumoral tissues. Moreover, lncRNA H19 could suppress the progression and metastasis of HCC. Citation32 Lv et al also reported that downregulation of lncRNA H19 and MiR-675 promotes migration and invasion of human HCC cells through AKT/GSK-3beta/Cdc25A signaling pathway. Citation33
lncRNA H19 has been recognized to act as an oncogenic driver in a number of cancer types, such as gastric, breast, and lung cancers. Citation34–Citation36 However, the specific function of lncRNA H19 in thyroid cancer remains controversial. For example, Wang et al reported that lncRNA H19 represses cell viability, and restrains migratory and invasive capabilities of thyroid cancer cells through downregulation of IRS-1. Citation37 Lan et al demonstrated that inhibition of lncRNA H19 promotes the proliferation and motility of PTC. Citation38 On the contrary, Liu et al reported that increased lncRNA H19 expression levels are associated with poor prognosis in thyroid cancer patients. Citation39 Another study by Liu et al suggested that increased expression of lncRNA H19 promoted growth, migration, and invasion in thyroid cancer, whereas H19 knockdown impaired TC cell viability. Citation40 Our analysis in the ONCOMINE database demonstrated that lncRNA H19 expression was markedly higher in thyroid cancer than normal tissue samples. Particularly, the qRT-PCR assay of 58 PTCs and corresponding adjacent nontumorous tissues from our institution also showed that lncRNA H19 expression was considerably higher in PTC tissues than paired paracancerous tissues. These findings consistently suggested that lncRNA H19 distinctively was expressed higher in PTCs than normal counterparts. Notably, we found that higher lncRNA H19 level was remarkably associated with larger tumor size, increased number of lymph node metastasis, as well as more advanced clinical stage, implying that higher lncRNA H19 level is correlated to higher tumor burden of PTCs.
Notably, emerging evidences in recent years indicated that lncRNA H19 acts as an important inducer of EMT. Citation12,Citation36,Citation41 Matouk et al have reported that the role of the oncofetal lncRNA H19 in tumor metastasis orchestrating the EMT-MET decision. Citation42 Zhou et al reported that the lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Citation43 However, whether lncRNA H19 acts as an important inducer of EMT in papillary thyroid cancer remained unclear. Our analysis with ONCOMINE and cBioPortal databases, as well as TCGA datasets consistently demonstrated that lncRNA H19 was positively associated with key EMT factors. It was found that higher expression of lncRNA H19 was correlated to elevated expression of Vimentin, as well as critical EMT-associated transcription factors, including ZEB2, Twist, and Snail2.
Loss or decreased E-cadherin and increased Vimentin are universally regarded as the hallmark of the EMT process. Citation44 In the present study, we first used the human PTC cell line B-CPAP to characterize the potential role of lncRNA H19 and found that lncRNA H19 was able to regulate mRNA and protein expression of EMT markers E-Cadherin and Vimentin, which suggested that lncRNA H19 was pivotal driver of EMT in PTC cells. In addition, our study also showed that lncRNA H19 promotes migration and invasion abilities of PTC cells. Shi et al reported that lncRNA H19 predicts poor prognosis in patients with melanoma and regulates cell growth, invasion, migration, and EMT in melanoma cells. Citation12 Liao et al reported that downregulation of lncRNA H19 inhibits the migration and invasion of melanoma cells by inactivating the NF-κB and PI3K/Akt signaling pathways. Citation45
Conclusion
In summary, the expression level of lncRNA H19 was significantly higher in PTC tissues than paired paracancerous tissue or normal tissues. Elevated lncRNA H19 expression was correlated with higher tumor burden of PTC. lncRNA H19 regulates mRNA and protein expression of classic EMT markers E-Cadherin and Vimentin and promotes migration and invasion of PTC cells. lncRNA H19 is a critical oncogenic driver in PTC, and targeted therapy against lncRNA H19 might be a potential avenue, in conjunction with other conventional therapy, to increase the efficacy for patients with PTC.
Acknowledgments
This work was partly supported by the Natural Science Foundation of Guangdong Province, China (No. 2018A030313562), National Natural Science Foundation of China (81600358), and Youth Innovative Talent Project of Colleges and Universities in Guangdong Province, China (No. 2017KQNCX073).
Supplementary material
Figure S1 Expression pattern of H19 in different tumor types.
Notes: This figure shows the number of datasets with statistically significant RNA overexpression (red) or down-expression (blue) of the target gene (cancer vs. normal tissue). The P-value threshold is 0.01. The number in each cell represents the number of analyses that meet the threshold within those analysis and cancer types. The gene rank was analyzed by percentile of target gene in the top of all genes measured in each research. Cell color is determined by the best gene rank percentile for the analyses within the cell.
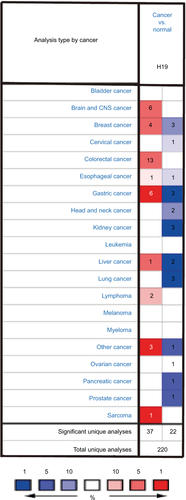
Figure S2 The RNA expression level of H19 in thyroid cancer cells ranked eleventh highest in a variety of cancer cell lines from Cancer Cell Line Encyclopedia analysis (shown in red frame).
Abbreviations: RMA, return material authorization; CML, chronic myelocytic leukemia; AML, acute myelocytic leukemia; NSC, non-small cell.
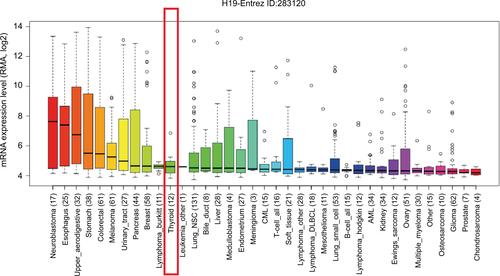
Table S1 Primers used in RT-PCR
Table S2 Oligonucleotide sequences for siRNA constructs
Table S3 Antibodies used in this study
Disclosure
The authors report no conflicts of interest in this work.
References
- FaginJAWellsSABiologic and clinical perspectives on thyroid cancerN Engl J Med2016375111054106727626519
- TengHMaoFLiangJTranscriptomic signature associated with carcinogenesis and aggressiveness of papillary thyroid carcinomaTheranostics20188164345435830214625
- ChenDQiWZhangPInvestigation of BRAF V600E detection approaches in papillary thyroid carcinomaPathol Res Pract2018214230330729254799
- KaiserMIEncode and the parts of the human genomeStud Hist Philos Biol Biomed Sci201872283730385203
- DavisCAHitzBCSloanCAThe encyclopedia of DNA elements (encode): data portal updateNucleic Acids Res201846D1D794D80129126249
- ThinKZLiuXFengXRaveendranSTuJCLncRNA-DANCR: a valuable cancer related long non-coding RNA for human cancersPathol Res Pract2018214680180529728310
- AchourCAguiloFLong non-coding RNA and polycomb: an intricate partnership in cancer biologyFront Biosci20182321062132
- Uszczynska-RatajczakBLagardeJFrankishAGuigóRJohnsonRTowards a complete map of the human long non-coding RNA transcriptomeNat Rev Genet201819953554829795125
- YoshimuraHMatsudaYYamamotoMKamiyaSIshiwataTExpression and role of long non-coding RNA H19 in carcinogenesisFront Biosci201823614625
- LiHYuBLiJOverexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancerOncotarget2014582318232924810858
- GuanG-FZhangD-JWenL-JOverexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinomaInt J Med Sci2016131291492227994496
- ShiGLiHGaoFTanQlncRNA H19 predicts poor prognosis in patients with melanoma and regulates cell growth, invasion, migration and epithelial-mesenchymal transition in melanoma cellsOnco Targets Ther2018113583359529950863
- ZhaoJMaS-TDownregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-κB pathwayMol Med Rep20181757388739429568924
- MitchellBDhingraJKMahalingamMBRAF and epithelial-mesenchymal transition: lessons from papillary thyroid carcinoma and primary cutaneous melanomaAdv Anat Pathol201623424427127145091
- KnaufJASartorMAMedvedovicMProgression of BRAF-induced thyroid cancer is associated with epithelial-mesenchymal transition requiring concomitant MAP kinase and TGFβ signalingOncogene201130283153316221383698
- BaqueroPSánchez-HernándezIJiménez-MoraEOrgazJLJiménezBChiloechesA(V600E)BRAF promotes invasiveness of thyroid cancer cells by decreasing E-cadherin expression through a Snail-dependent mechanismCancer Lett2013335123224123435375
- KimTHKimYNKimHIPrognostic value of the eighth edition AJCC TNM classification for differentiated thyroid carcinomaOral Oncol201771818628688696
- PaulinCFabienNFuscoADescription of cell line established from human thyroid papillary cancer and secreting human chorionic gonadotropin hormoneC R Acad Sci III199231512493498 French1297527
- RhodesDRKalyana-SundaramSMahavisnoVOncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profilesNeoplasia20079216618017356713
- GaoJAksoyBADogrusozUIntegrative analysis of complex cancer genomics and clinical profiles using the cBioPortalSci Signal20136269pl123550210
- LinH-YZengDLiangY-KWeiX-LChenC-FGATA3 and TRPS1 are distinct biomarkers and prognostic factors in breast cancer: database mining for GATA family members in malignanciesOncotarget2017821347503476128423734
- ZengDXiaoYZhuJPengCLiangWLinHKnockdown of nucleophosmin 1 suppresses proliferation of triple-negative breast cancer cells through activating CDH1/Skp2/p27kip1 pathwayCancer Manag Res20191114315630613163
- ShaukatAAThe rising trend in papillary thyroid carcinoma. True increase or over diagnosis?Saudi Med J201839553129738019
- JillardCLScheriRPSosaJAWhat is the optimal treatment of papillary thyroid cancer?Adv Surg201549799326299491
- QuagliarielloVArmeniaEAurilioCNew treatment of medullary and papillary human thyroid cancer: biological effects of hyaluronic acid hydrogel loaded with quercetin alone or in combination to an inhibitor of Aurora kinaseJ Cell Physiol201623181784179526660542
- HuangYQuSZhuGBRAF V600E Mutation-Assisted risk stratification of solitary Intrathyroidal papillary thyroid cancer for precision treatmentJ Natl Cancer Inst2018110436237029165667
- KwongNMarquseeEGordonMSLong-term, treatment-free survival in select patients with distant metastatic papillary thyroid cancerEndocr Connect20143420721425316293
- PerezDSHoageTRPritchettJRLong, abundantly expressed non-coding transcripts are altered in cancerHum Mol Genet200817564265518006640
- ErhoNBuerkiCTricheTJDavicioniEVergaraIATranscriptome-wide detection of differentially expressed coding and non-coding transcripts and their clinical significance in prostate cancerJ Oncol201220124111
- VuTNPramanaSCalzaSSuoCLeeDPawitanYComprehensive landscape of subtype-specific coding and non-coding RNA transcripts in breast cancerOncotarget2016742688516886327634900
- ZhangTWangY-RZengFCaoH-YZhouH-DWangY-JLncRNA H19 is overexpressed in glioma tissue, is negatively associated with patient survival, and promotes tumor growth through its derivative miR-675Eur Rev Med Pharmacol Sci201620234891489727981546
- ZhangLYangFYuanJ-HangEpigenetic activation of the miR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinomaCarcinogenesis201334357758623222811
- LvJMaLChenX-LHuangX-HWangQDownregulation of LncRNAH19 and miR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3β/Cdc25A signaling pathwayJ Huazhong Univ Sci Technolog Med Sci201434336336924939300
- YörükerEEKeskinMKulleCBHoldenriederSGezerUDiagnostic and prognostic value of circulating lncRNA H19 in gastric cancerBiomed Rep20189218118630083318
- SunHWangGPengYH19 lncRNA mediates 17β-estradiol-induced cell proliferation in MCF-7 breast cancer cellsOncol Rep20153363045305225846769
- ZhangQLiXLiXLiXChenZLncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cellsJ Cell Biochem201811964447445729219208
- WangPLiuGXuWLiuHBuQSunDLong noncoding RNA H19 inhibits cell viability, migration, and invasion via downregulation of IRS-1 in thyroid cancer cellsTechnol Cancer Res Treat20171661102111229332545
- LanXSunWDongWDownregulation of long noncoding RNA H19 contributes to the proliferation and migration of papillary thyroid carcinomaGene20186469810529287713
- LiuNZhouQQiY-HWangHYangLFanQ-YEffects of long non-coding RNA H19 and microRNA let7a expression on thyroid cancer prognosisExp Mol Pathol20171031717728655518
- LiuLYangJZhuXLiDLvZZhangXLong noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancerFebs J2016283122326233927093644
- JiaLTianYChenYZhangGThe silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/β-catenin pathwayOnco Targets Ther20181131332129391808
- MatoukIJHalleDRavehEGilonMSorinVHochbergAThe role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decisionOncotarget2016743748376526623562
- ZhouWYeX-LXuJThe lncRNA H19 mediates breast cancer cell plasticity during EMT and Met plasticity by differentially sponging miR-200b/c and let-7bSci Signal201710483eaak955713
- VoonDCHuangRYJacksonRAThieryJPThe EMT spectrum and therapeutic opportunitiesMol Oncol201711787889128544151
- LiaoZZhaoJYangYDownregulation of lncRNA H19 inhibits the migration and invasion of melanoma cells by inactivating the NFκB and PI3K/Akt signaling pathwaysMol Med Rep20181757313731829568965
