Abstract
Background
Abnormal expression of long non-coding RNA anti-differentiation noncoding RNA (lncRNA DANCR) can frequently be detected in cancer. Because of this, it is of vital necessity to perform a meta-analysis to clarify the value of lncRNA DANCR as a prognostic marker in malignant tumors.
Methods
Related studies were retrieved from electronic databases including Web of Science, PubMed, and OVID, from inception to November 21, 2018. The HRs and corresponding 95% CIs were also calculated to explore the relationship of lncRNA DANCR expression with patient survival. Moreover, ORs were computed to assess the association of lncRNA DANCR expression with the pathological parameters.
Results
A total of 14 studies involving 1,117 patients were included in this meta-analysis. The pooled HR suggested that high lncRNA DANCR expression was correlated with poor overall survival (OS; HR =1.85, 95% CI: 1.56–2.18) and disease-free survival (DFS; HR =2.49, 95% CI: 1.75–3.56) in cancer patients. Besides, High lncRNA DANCR expression was related to poor histological grade (PHG; OR =2.01, 95% CI: 1.08–3.75), high tumor stage (HTS; OR =3.52, 95% CI: 1.67–7.43), lymph node metastasis (LNM; OR =3.47, 95% CI: 1.42–8.49), and distant metastasis (DM; OR =4.76, 95% CI: 2.39–9.51). However, no evidence of obvious asymmetry was found for DFS (Pr>|z|=0.308), PHG (Pr>|z|=0.707), LNM (Pr>|z|=0.174), and DM (Pr>|z|=0.734) using Begg’s funnel plot.
Conclusion
Our findings suggest that high lncRNA DANCR expression can predict poor OS, DFS, PHG, HTS, LNM, and DM in cancer patients, implying that high lncRNA DANCR expression may potentially serve as a new indicator for poor prognosis and metastasis in cancer.
Keywords:
Introduction
Recent report demonstrates that, the US has witnessed about 1.7 million new cancer cases and 600,000 cancer-related deaths in 2017. Citation1 Nevertheless, the 5-year survival of most cancers remains dismally low, and a large number of scientists are devoting themselves to looking for new biomarkers to determine or diagnose cancer prognosis.
lncRNA, which lacks a meaningful open reading frame, is defined as the transcribed RNA molecule that is >200 nucleotides in length, which has possessed many important functions in disease, such as posttranscriptional, transcriptional, and epigenetic regulation. Citation2,Citation3 In addition, abnormal lncRNA expression is currently recognized to be related to various cancer types. Citation4–Citation7 For instance, some lncRNAs play crucial parts in metastasis, invasion, and proliferation of cancer cells, indicating that lncRNA may serve be a useful marker for predicting cancer prognosis. Citation8–Citation10
Typically, the lncRNA DANCR was discovered by Kretz et al in 2012, which was originally deemed to be essential for the dedifferentiation of epidermal cells. Citation11 Besides, recent studies reveal that DANCR plays a crucial role in the differentiation of periodontal ligament stem cells into osteoblasts, which can also promote tumor cell dissemination and metastasis formation. Citation12–Citation14 Moreover, lncRNA DANCR is also suggested in some studies to be correlated with different tumor biological parameters, such as tumor growth, metastasis, and progression. Citation15–Citation17 Metastasis and prognosis may be affected by lncRNA DANCR; nonetheless, a majority of existing studies are limited by their small sample sizes and discrete outcomes. As a consequence, an updated meta-analysis was performed in this study to determine the prognostic value of lncRNA DANCR in cancer patients.
Materials and methods
Literature collection
In accordance with the standard guidelines for meta-analyses,Citation18,Citation19 related articles that served lncRNA DANCR as a prognostic biomarker for the survival of cancer patients were systemically retrieved from some online databases by two authors independently from inception to November 21, 2018. Meanwhile, text words and Mesh strategies were adjusted based on the databases in this retrieval, including the following terms (“Long non-coding RNA differentiation antagonizing non-protein coding RNA“ or “lncRNA DANCR” or “lncRNA ANCR”) and (“recurrence” or “outcome” or “survival”, “cancer” or “neoplasm” or “tumor” or “carcinoma”, “prognosis” or “prognostic”). Moreover, the reference lists of relevant articles were also manually retrieved during retrieval, so as to avoid missing any potentially eligible studies.
Study selection
All the included studies were then evaluated, and data were extracted by two scholars independently. Typically, the study inclusion criteria were as follows: 1) studies in which all tumors were confirmed by histological or pathological examinations; 2) studies in which the lncRNA DANCR expression levels in human tumor tissues were measured; 3) studies in which patients were grouped in accordance with different lncRNA DANCR expression levels, and the cutoff values of high and low DANCR expression might be the median or mean of all samples in their study; and 4) studies with sufficient original data for statistical analyses of pathological or patient survival parameters with lncRNA DANCR expression.
In addition, the study exclusion criteria were shown below: non-human studies and non-English studies; editorials, reviews, expert opinions as well as letters; database analysis without original data; and studies mentioning functions and molecular structure of lncRNA DANCR only.
Date extraction
Data from the original articles were independently examined and extracted by two reviewers, and any disagreement between them during the process of literature assessment was settled by the consensus with a third reviewer. A series of data were collected in this meta-analysis, including surname of the first author, publication year, country, tumor type, sample size, number of patients with LTS, PHG, HTS, LNM and DM, reference gene and detection method of lncRNA DANCR, as well as HRs and 95% CIs of elevated lncRNA DANCR expression for OS and DFS.
Statistical methods
The Stata version 12.0 software was adopted for all statistical analyses. In addition, the heterogeneity was also measured in this meta-analysis using Q and I2 tests. The test results had indicated the presence of significant heterogeneity in this research (I2≥50%, and P<0.1);Citation20 therefore, the random effect model should be adopted. Besides, the potential publication bias was also assessed by Egger’s test and Begg’s funnel plot. The pooled ORs and HRs should be extracted from the published data; typically, the crude data should be adopted if the HRs could not be obtained directly from the publications. Besides, the survival information extracted from Kaplan–Meier curves should be adopted to estimate the HRs when they were not directly reported in the studies. To make a summary about the outcomes of survival, both SE and the log HR should be collected. Citation21 Moreover, 95% CIs and ORs should be combined to assess the relationship of clinicopathological parameters with lncRNA DANCR.
Results
Study characteristics
Details about the screening process are shown in . In accordance with the exclusion and inclusion criteria, 14 studies involving 1,117 patients were enrolled into this meta-analysis. Citation22–Citation35 Characteristics of the 14 studies included in this meta-analysis are summarized in . As could be observed, the sample size in the 14 studies ranged from 34 to 135, with an average of 79.57. Besides, all the enrolled studies were published between 2015 and 2018 and were carried out in China. Among these studies, respectively, one study had focused on CVR,Citation25 TNBC,Citation29 RB,Citation30 HCC,Citation34 and BC;Citation35 three concentrated on GC;Citation22,Citation27,Citation28 two focused on OSC;Citation23,Citation32 two on glioma;Citation24,Citation33 and two on CRC. Citation26,Citation31 All clinical pathological parameters were dependent on the pathology. Moreover, it was found that the reference genes of lncRNA DANCR were different among these studies, which had included GAPDH,Citation23–Citation27,Citation29–Citation34 β-actin,Citation22,Citation35 and small nuclear RNA U6. Citation28 Moreover, the thresholds of high and low lncRNA DANCR expression levels, including the median and average lncRNA DANCR expression, were also different among these studies.
Table 1 The basic information and data of all included studies in the meta-analysis
Association between the lncRNA DANCR expression level and survival
To assess the role of lncRNA DANCR in OS for cancer patients, cumulative meta-analysis was carried out in this research. As shown, the relationship of OS with lncRNA DANCR was reported in ten studies enrolling 839 patients (). Meanwhile, the fixed effects model was adopted since there was no significant heterogeneity (I2=0.0%, PQ=0.728). The results suggested that the OS in cancer patients was markedly related to the lncRNA DANCR expression (pooled HR =1.85, 95% CI: 1.56–2.18; ). Besides, sensitivity analysis was also carried out, which had confirmed the robustness of these results (). Subsequently, subgroup analyses stratified by cancer type, sample size, NOS score, and HR statistic method were also carried out (, ).
Figure 2 Forest plot (A) and sensitivity analysis (B) showed the relationship between lncRNA DANCR expression level and OS in cancer.
Abbreviations: lncRNA DANCR, long non-coding RNA anti-differentiation noncoding RNA; OS, overall survival.
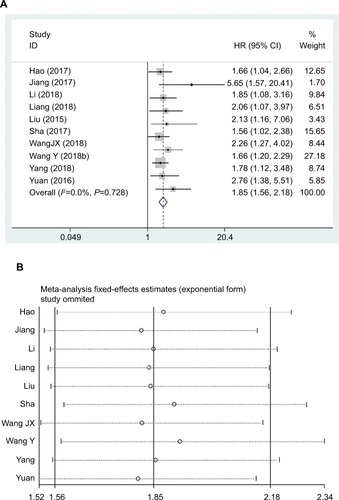
Figure 3 Forest plots of subgroup analysis for OS of patients with cancer.
Notes: Subgroup analysis by tumor type (A), sample size (B), NOS score (C), and HR statistic method (D).
Abbreviations: NOS, Newcastle–Ottawa Scale; OS, overall survival.

Table 2 Survival data of studies included in the meta-analysis
Table 3 Subgroup analysis of OS by tumor type, sample size, NOS score, and HR statistic method
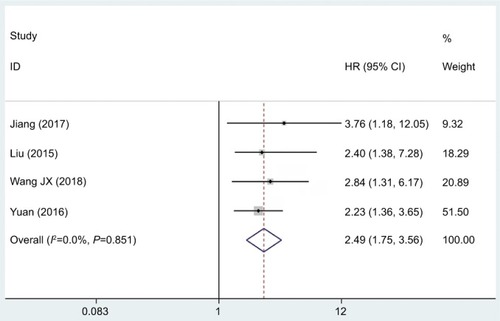
Moreover, cumulative meta-analysis was also performed to determine the role of lncRNA DANCR in DFS among the 330 cancer patients recruited into the eligible studies (). The results revealed that lncRNA DANCR was correlated with DFS (pooled HR =2.49, 95% CI: 1.75–3.56) in cancer patients upon statistical analyses. Similarly, the fixed effects model was employed due to the insignificant heterogeneity.
Figure 4 Forest plot showed the relationship between lncRNA DANCR expression level and DFS in cancer.
Abbreviations: DFS, disease-free survival; lncRNA DANCR, long non-coding RNA anti-differentiation noncoding RNA.
These results suggested that the shorter OS and DFS in cancer patients might be associated with higher lncRNA DANCR expression. As a result, it could be concluded that lncRNA DANCR was an independent factor of the survival for cancer patients.
Association between the lncRNA DANCR expression level and LTS
shows the association between LTS and lncRNA DANCR expression from ten studies involving 757 patients. Specifically, the random-effects model was adopted due to the presence of a significant heterogeneity among the eligible studies (I2=79.4%, PQ=0.000). Our results had revealed a pooled OR of 1.63 (95% CI: 0.80–3.31; high vs low lncRNA DANCR expression). Moreover, sensitivity analysis of all included studies was also performed, and the OR of high to low expression groups was 2.10 (95% CI: 1.25–3.54) after the study by Hao et alCitation22 was excluded (I2=56.6%, PQ=0.018) ( and ).
Figure 5 Forest plot (A), sensitivity analysis (B), and the forest plot of sensitivity analysis (C) showed the association between LTS and lncRNA DANCR expression level in cancer.
Abbreviations: lncRNA DANCR, long non-coding RNA anti-differentiation noncoding RNA; LTS, larger tumor size.
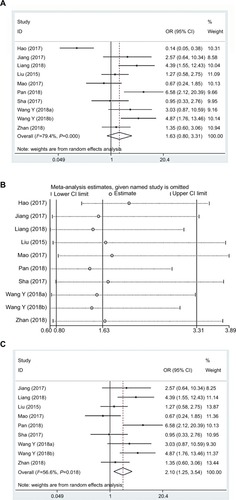
Conforming to the abovementioned results, no significant difference was detected in the LTS incidence between two groups, but additional studies were needed to confirm the association between lncRNA DANCR and LTS in cancer patients.
Association between the lncRNA DANCR expression level and PHG
In this research, data regarding the association between the lncRNA DANCR expression and PHG had been collected from six eligible studies involving 503 cancer patients, and the random-effects model was adopted as a result of the significant heterogeneity (I2=61.4%, PQ=0.024). Besides, the OR of high to low lncRNA DANCR expression groups was 2.10 (95% CI: 1.08–3.75, ). Typically, the heterogeneity had disappeared (I2=24.2%, PQ=0.266) after two studies were removed in sensitivity analysis, with the OR of high to low expression groups of 3.14 (95% CI: 1.95–5.05) ( and ).
Figure 6 Forest plot (A), sensitivity analysis (B), and the forest plot of sensitivity analysis (C) showed the association between PHG and lncRNA DANCR expression level in cancer.
Abbreviations: lncRNA DANCR, long non-coding RNA anti-differentiation noncoding RNA; PHG, poor histological grade.
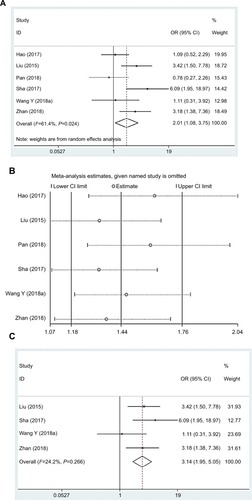
In accordance with these results, a significant difference was noted in the incidence of PHG between two groups, indicating that the risk of PHG was remarkably correlated with high lncRNA DANCR expression.
Association between the lncRNA DANCR expression level and HTS
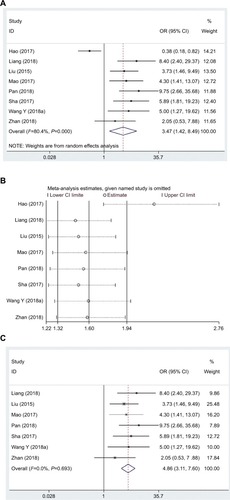
In this meta-analysis, the correlation between HTS and lncRNA DANCR expression was detected in ten eligible studies recruiting 809 patients. Similarly, the random effects model would be adopted (I2=81.4%, PQ=0.000). The results discovered that HTS in cancer patients was notably related to high lncRNA DANCR expression (pooled OR =3.52, 95% CI: 1.67–7.43, ). In addition, the heterogeneity had disappeared in sensitivity analysis after the study by Hao et alCitation22 was excluded (I2=0.0%, PQ=0.905), and the OR of high to low lncRNA DANCR expression groups was 4.67 (95% CI: 3.30–6.60) ( and ).
Figure 7 Forest plot (A), sensitivity analysis (B), and the forest plot of sensitivity analysis (C) showed the association between HTS and lncRNA DANCR expression level in cancer.
Abbreviations: HTS, high tumor stage; lncRNA DANCR, long non-coding RNA anti-differentiation noncoding RNA.
According to the analysis results, compared with the low lncRNA DANCR expression group, the tumor stage in high lncRNA DANCR expression group was markedly higher, demonstrating that the risk of HTS was evidently correlated with high lncRNA DANCR expression.
Association between the lncRNA DANCR expression level and LNM
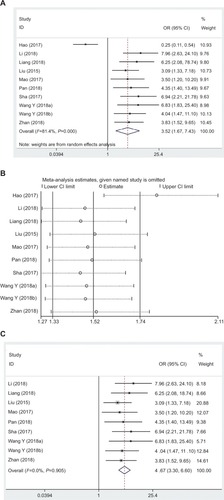
In this research, data collected from eight eligible studies involving 628 cancer patients were also analyzed, and the random effects model had been adopted based on the significant heterogeneity (I2=80.4%, PQ=0.000). Additionally, the OR of to low lncRNA DANCR expression groups was 3.47 (95% CI: 1.42–8.49, ). Consistent with the results of previous sensitivity analysis, the heterogeneity had disappeared (I2=0.0%, PQ=0.693) after the study by Hao et alCitation22 was removed ( and ).
Figure 8 Forest plot (A), sensitivity analysis (B), and the forest plot of sensitivity analysis (C) showed the association between LNM and lncRNA DANCR expression level in cancer.
Abbreviations: lncRNA DANCR, long non-coding RNA anti-differentiation noncoding RNA; LNM, lymph node metastasis.
In accordance with these results, a significant difference was noted between two groups in terms of LNM incidence. As far as cancer patients were concerned, high lncRNA DANCR expression was markedly correlated with greater susceptibility to LNM.
Association between the lncRNA DANCR expression level and DM
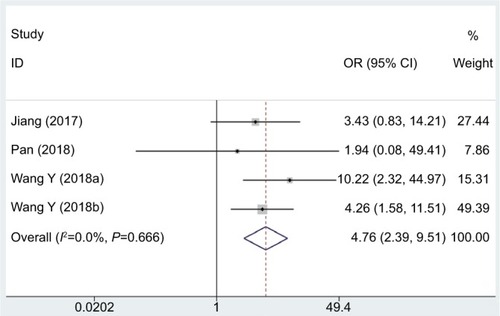
In this meta-analysis, the correlation of DM with the lncRNA DANCR expression level was examined in four eligible studies including 241 patients, and the fix effects model was adopted due to the limited heterogeneity (I2=0.0%, PQ=0.666). The OR of high to low lncRNA DANCR expression groups was 4.76 (95% CI: 2.39–9.51, ). Consistent with these results, the DM incidence was significantly different between two groups, revealing that high lncRNA DANCR expression could remarkably predict a higher tendency to develop DM in cancer patients.
Publication bias
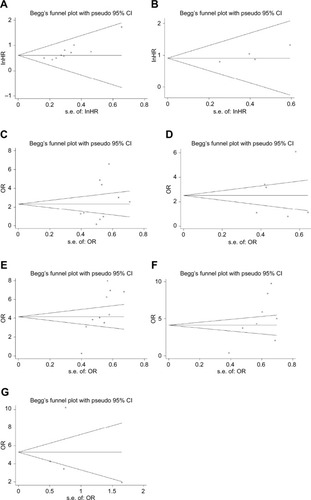
Subsequently, the Begg’s funnel plot was conducted in this study to evaluate the potential publication bias. shows no evidence of obvious asymmetry for DFS (Pr>|z|=0.308), LTS (Pr>|z|=0.283), PHG (Pr>|z|=0.707), LNM (Pr>|z|=0.174), and DM (Pr>|z|=0.734). However, significant publication bias was detected for OS (Pr>|z|=0.004) and HTS (Pr>|z|=0.007).
Figure 10 Begg’s publication bias plots evaluating the relationship between lncRNA DANCR expression and OS (A), DFS (B), LTS (C), PHG (D), HTS (E), LNM (F), DM (G).
Abbreviations: DFS, disease-free survival; DM, distant metastasis; HTS, high tumor stage; lncRNA DANCR, long non-coding RNA anti-differentiation noncoding RNA; LNM, lymph node metastasis; LTS, larger tumor size; OS, overall survival; PHG, poor histological grade.
Discussion
Cancer still poses a serious threat to human health, which is gradually increased in recent years in terms of morbidity. Citation1 Nonetheless, the exact metastasis mechanism in cancer patients remains unclear despite that metastasis is an important indicator of poor prognosis. Citation36,Citation37 Therefore, it is necessary to identify new molecular markers to predict tumor metastasis at present, since they may play critical roles in treating and predicting cancer. Citation38 lncRNAs, one of these molecular markers, can affect tumor initiation, progression, and occurrence, which can easily collect the useful biomarkers for cancer monitoring and diagnosis. Citation39–Citation41
lncRNA DANCR has been verified in previous studies to be an important oncogene in various human cancers, including GC, glioma, CVR, OSC, CRC, RB, HCC, and BC. Citation22–Citation35 Additionally, lncRNA DANCR expression has been confirmed in recent study to be upregulated in CRC tissues, which is correlated with poor survival for CRC patients. Citation26,Citation31 Moreover, according to Li et al, DANCR could positively promote the proliferation and migration of glioma through activating the Wnt/β-catenin signaling pathway. Citation24 Besides, Mao et al also reported that DANCR was upregulated in GC tissues, which could enhance the migration and invasion of GC cells. Citation27 Additionally, Wang et al found that DANCR could strongly suppress HCC proliferation via targeting miR-216a-5p and KLF12. Citation42 Furthermore, Lu et al demonstrated that DANCR was elevated in a broad spectrum of human cancers, and MYC could drive cancer cell proliferation by targeting DANCR. Citation43 These results reveal that lncRNA DANCR may be a crucial prognostic factor for cancer patients. Nevertheless, the underlying mechanisms by which lncRNA DANCR affects cancer remain unknown so far. Therefore, this meta-analysis was performed to examine the prognostic value and clinicopathological significance of lncRNA DANCR in cancer patients.
In this research, related data collected from the 14 eligible studies involving 1,117 cancer patients were analyzed, and a fixed or a random effects model had been adopted based on the heterogeneity analysis results. For cancer patients, high lncRNA DANCR expression could potentially serve as an indicator of poor prognosis. Besides, significant differences were found in OS and DFS between the two groups after combining HRs from the Cox multivariate analyses, and it was found that poor OS and DFS in various cancer kinds were associated with high lncRNA DANCR expression. Moreover, high lncRNA DANCR expression in cancer patients was also remarkably related to some clinicopathological parameters, including PHG, HTS, DM, and LNM. To sum up, findings of this meta-analysis indicated that lncRNA DANCR might serve as a valuable biomarker for the poor prognosis of most cancers.
Limitations
Several limitations should be taken into consideration when interpreting the conclusion of this meta-analysis. First, data in this meta-analysis might not be applicable for countries all over the world, since all the included studies were from China. Second, in spite of the best effort made to search for all relevant studies only 14 studies were ultimately enrolled in this study; the relatively small sample size might reduce the stringency of our conclusion. Third, the criterion of high expression was not consistent among all articles, making it difficult to obtain the same value. Last but not least, there were other factors that might affect cancer prognosis, such as comorbidities and therapies, but related information was not available in the analyzed enrolled articles, which had therefore become an inherent shortcoming of this systematic review and meta-analysis. As a consequence, the role of lncRNA DANCR in cancer should be further confirmed by more high-quality and well-designed studies.
Conclusion
To sum up, our findings suggest that high lncRNA DANCR expression in a series of cancers is remarkably correlated with poor OS, DFS, PHG, HTS, DM, and LNM. As a result, lncRNA DANCR may potentially serve as a biomarker to determine metastasis and predict the prognosis for cancer patients.
Abbreviations
| BC | = | bladder cancer |
| CRC | = | colorectal cancer |
| CVR | = | cervical cancer |
| DFS | = | disease-free survival |
| DM | = | distant metastasis |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| GC | = | gastric cancer |
| HCC | = | hepatocellular carcinoma |
| HTS | = | high tumor stage |
| lncRNA DANCR | = | long non-coding RNA anti-differentiation noncoding RNA |
| LNM | = | lymph node metastasis |
| LTS | = | larger tumor size |
| NOS | = | Newcastle–Ottawa Scale |
| OS | = | overall survival |
| OSC | = | osteosarcoma |
| PHG | = | poor histological grade |
| RB | = | retinoblastoma |
| TNBC | = | triple negative breast cancer. |
Acknowledgments
This article was supported by the Science and Technology Development Program of Shandong Province (No. 2015GSF118110).
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRLMillerKDJemalACancer statistics, 2017CA Cancer J Clin201767173028055103
- JohnssonPLipovichLGranderDMorrisKVEvolutionary conservation of long non-coding RNAs; sequence, structure, functionBiochim Biophys Acta18402014310631071
- FanYHFangHJiCXXieHXiaoBZhuXGLong noncoding RNA CCAT2 can predict metastasis and poor prognosis: a meta-analysisClin Chim Acta201746612012628089750
- DaiWTianCJinSEffect of lncRNA ANRIL silencing on anoikis and cell cycle in human glioma via microRNA-203aOnco Targets Ther2018115103510930197521
- ZhengYGaoYLiXLong non-coding RNA NAP1L6 promotes tumor progression and predicts poor prognosis in prostate cancer by targeting inhibin-β AOnco Targets Ther2018114965497730154665
- LiuQLiuHChengHLiYLiXZhuCDownregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cellsOnco Targets Ther2017102461247128503069
- HuaJTAhmedMGuoHRisk SNP-Mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19Cell20181743564575.e1830033362
- MaZHuangHXuYCurrent advances of long non-coding RNA highly upregulated in liver cancer in human tumorsOnco Targets Ther2017104711471729026319
- FanYYanTChaiYJiangYZhuXLong noncoding RNA HOTTIP as an independent prognostic marker in cancerClinica Chimica Acta2018482224230
- GuoHAhmedMZhangFModulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancerNat Genet201648101142115027526323
- KretzMWebsterDEFlockhartRJSuppression of progenitor differentiation requires the long noncoding RNA ANCRGenes Dev201226433834322302877
- JiaQJiangWNiLDown-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cellsArch Oral Biol201560223424125463901
- ZhuLXuPCDownregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expressionBiochem Biophys Res Commun2013432461261723438432
- JiaJLiFTangXSLong noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3Oncotarget2016725378683788127191265
- MaXWangXINYangCDANCR acts as a diagnostic biomarker and promotes tumor growth and metastasis in hepatocellular carcinomaAnticancer Res201636126389639827919960
- ZhenQGaoLNWangRFLncRNA DANCR promotes lung cancer by sequestering miR-216aCancer Control2018251107327481876984
- YuXu DGaoJLuGZhangGMaY PLncRNA DANCR functions as a competing endogenous RNA to regulate Rab1a expression by sponging miR-634 in gliomaBiosci Rep2018381BSR2017166429301870
- AltmanDGMcShaneLMSauerbreiWTaubeSEReporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaborationPLoS Med201295e100121622675273
- HarrisALReporting recommendations for tumour MARKer prognostic studies (REMARK)Br J Cancer200593438538616106244
- FanYHJiCXXuBFanHYChengZJZhuXGLong noncoding RNA activated by TGF-beta in human cancers: a meta-analysisClin Chim Acta2017468101628163033
- MaPJGuanQKXuDWZhaoJQinNJinBZLncRNA PANDAR as a prognostic marker in Chinese cancerClin Chim Acta201747517217729066211
- HaoYPQiuJHZhangDBYuCGLong non-coding RNA DANCR, a prognostic indicator, promotes cell growth and tumorigenicity in gastric cancerTumour Biol2017396101042831769979828618943
- JiangNWangXXieXlncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating Axl via miR-33a-5p inhibitionCancer Lett2017405465528642170
- LiJZhouLOverexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/β-catenin signalingBiomed Pharmacother201810260260729602127
- LiangHZhangCGuanHLiuJCuiYLncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5pJ Cell Physiol201923457266727830362591
- LiuYZhangMLiangLLiJChenYXOver-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancerInt J Clin Exp Pathol201589114801148426617879
- MaoZLiHDuBLncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cellsBiosci Rep2017376BSR2017107028951520
- PanLLiangWGuJLong noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cellsOncotarget2018921915193029416741
- ShaSYuanDLiuYHanBZhongNTargeting long non-coding RNA DANCR inhibits triple negative breast cancer progressionBiol Open2017691310131628760736
- WangJXYangYLiKLong noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9J Cell Physiol2018233106986699529744877
- WangYLuZWangNLong noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 spongingExp Mol Med20185055729717105
- WangYZengXWangNLong noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcomaMol Cancer20181718929753317
- YangJXSunYGaoLMengQYangBYLong non-coding RNA DANCR facilitates glioma malignancy by sponging miR-33a-5pNeoplasma201865579079829940760
- YuanSXWangJYangFLong noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1Hepatology201663249951125964079
- ZhanYChenZLiYLong non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNAJ Exp Clin Cancer Res201837127330419948
- GuanHMeiYMiYDownregulation of lncRNA ANRIL suppresses growth and metastasis in human osteosarcoma cellsOnco Targets Ther2018114893489930147340
- LiuHPanYHanXLiuJLiRMicroRNA-216a promotes the metastasis and epithelial-mesenchymal transition of ovarian cancer by suppressing the PTEN/Akt pathwayOnco Targets Ther2017102701270928579808
- FanYHYeMHWuLOverexpression of miR-98 inhibits cell invasion in glioma cell lines via downregulation of IKKεEur Rev Med Pharmacol Sci201519193593360426502849
- QiPDuXThe long non-coding RNAs, a new cancer diagnostic and therapeutic gold mineMod Pathol201326215516522996375
- GaoPWeiGHGenomic insight into the role of lncRNA in cancer susceptibilityInt J Mol Sci2017186E123928598379
- GaoPXiaJHSipekyCBiology and clinical implications of the 19q13 aggressive prostate cancer susceptibility locusCell20181743576589.e1830033361
- WangJPuJZhangYDANCR contributed to hepatocellular carcinoma malignancy via sponging miR-216a-5p and modulating KLF12J Cell Physiol201923469408941630430564
- LuYHuZMangalaLSMyc targeted long noncoding RNA DANCR promotes cancer in part by reducing p21 levelsCancer Res2018781647429180471