Abstract
Background:
miR-101 is reported to be associated with cell proliferation and apoptosis. However, it is unknown whether miR-101 expression affects cell proliferation and apoptosis in diffuse large B cell lymphoma (DLBCL). The aim of the present study was to investigate the expression of miR-101 and its effect on cell proliferation and apoptosis in DLBCL.
Methods:
miR-101 expression was detected in 30 cases of patients with DLBCL and normal lymph node by qRT-PCR. Then, miR-101 expression was up-regulated and down-regulated in Originated Cell Line-Large Lymphoma 8 (OCL-LY8) cell line, respectively. MTT and flow cytometry assay were used to evaluate the effect of miR-101 on cell proliferation and apoptosis, respectively. As KDM1A was confirmed to be as a specific target of miR-101 by TargetScanHuman, the relationship between MiR-101 and KDM1A was further investigated.
Results:
miR-101 expression in patients with DLBCL was significantly reduced compared those in normal lymph node (P<0.05). miR-101 expression was significantly associated with tumor size, clinical stage and International Prognostic Index (IPI) scores (P<0.05). In OCL-LY8 cell line, miR-101 down-regulation significantly promoted cell proliferation and suppressed cell apoptosis. Meanwhile, miR-101 up-regulation reversed this effect. In addition, miR-101 negatively regulated the expression of KDM1A. KDM1A down-regulation was oberved in normal tissues compared with those in DLBCL tissues, which inhibited cell proliferation and promoted cell apoptosis.
Conclusion:
These data indicate that miR-101 regulates cell proliferation and apoptosis by targeting KDM1A, which provides a potential therapeutic for DLBCL patients.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma and is regarded as the most aggressive lymphoma worldwide.Citation1,Citation2 As early-stage DLBCL is hard to be diagnosed, DLBCL patients present poor prognosis.Citation3,Citation4 It is reported that 30%–40% of DLBCL patients might be refractory or relapse, even if they received a better therapeutic response to the first-line chemo-immunotherapy.Citation5 Currently, the molecular mechanism of DLBCL formation and development remains unclear. Therefore, identifying the biomarkers associated with DLBCL has become more urgent.
miRNA is a kind of sn-cRNA that regulates gene expression at the translational and post-transcriptional levels.Citation6,Citation7 Current reports indicate that miRNA plays a key role in tumor formation and development by regulating cell apoptosis, proliferation, invasion and migration.Citation8,Citation9 miR-101 is a highly conserved miRNA, which is located in chromosome1p31.3 and 9p24.1.Citation10 miR-101, as a tumor-suppressive miRNA, is lowly expressed in several cancer types, such as gastric cancer,Citation11 hepatocellular carcinoma,Citation12 melanomaCitation13 and esophageal cancer.Citation14 Moreover, miR-101 is involved in cell apoptosis, proliferation, migration and invasion.Citation14–Citation17 However, the expression and biological functions of miR-101 in DLBCL are unknown.
In this study, the expression and biological functions of miR-101 were investigated in DLBCL. The potential molecular mechanism of miR-101 in cell apoptosis and proliferation was explored.
Methods
Tissue sample collection and preparation
This study enrolled 30 lymph node samples from patients diagnosed with DLBCL and 30 individuals with normal lymph nodes as controls (NC). All samples were obtained from the Department of Hematology, Zhangzhou Affiliated Hospital of Fujian Medical University, People’s Republic of China, between July 2015 and December 2017. No patient received preoperative radiotherapy or chemotherapy before surgery. Samples were snap-frozen and stored in liquid nitrogen. All DLBCL diagnoses were confirmed by the pathologist on the basis of morphology and immunophenotypic findings. Clinicopathological features including age, gender, tumor size, extra-nodal status, B symptoms, lactate dehydrogenase (LDH), clinical stage and International Prognostic Index (IPI) score were collected from medical records and pathology reports. This study was conducted in accordance with the Declaration of Helsinki and supported by the Research Ethics Committee of Zhangzhou Affiliated Hospital of Fujian Medical University. All participants signed the informed consents.
Cell culture and transfection
The OCL-LY7, OCL-LY8 and OCL-LY10 cell lines were obtained from American Type Culture Collection (Manassas, VA, USA). All cells were cultured in RPMI-1640 containing 12% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a humidified chamber supplemented with 5% CO2. Cell transfection was performed according to the manufacturer’s protocol. OCL-LY8 cells were selected for the down-regulation and up-regulation experiment by transferring miR-101 siRNA/mimics (GenePharma, Suzhou, People’s Republic of China), using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.). Scramble siRNA served as a control. The siRNA sequence for KDM1A was 5′-GAGGAAGAGGAAAUGGCUG-3′.
qRT-PCR
Total RNA was extracted from tissue and cell specimens using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s protocol. The detection of qRT-PCR was performed by using SYBR green qPCR assay (Takara, People’s Republic of China). The primers of miR-101 and U6 were purchased from Riobio Biotech Corporation (Guangzhou, People’s Republic of China). The primers of KDM1A were 5′-CTGATGCAGGCCATCAAGT-3′ (forward) and 5′-TCTCCAGGAAATGCATTGGT-3′ (reverse). The primers of GAPDH were 5′-TGGTGGACCTCATGGCCTAC-3′ (forward) and 5′-CAGCAACTGAGGGCCTCTCT-3′ (reverse). PCR reaction condition was as follow: 95°C for 15 minutes, followed by 40 cycles of 94°C for 15 seconds, 60°C for 30 seconds and 70°C for 30 seconds. Relative expression level was compared by the 2−∆∆CT method.
Western blotting
Western blotting was used to detect KDM1A protein expression in OCL-LY8 cells. At 48 hours following transfection, total protein was extracted from cell specimens using lysis buffer containing 1% phenylmethanesulfonyl fluoride. Protein concentration was measured by bicinchoninic acid kit. Then, 50 µg protein specimens were separated by 10% SDS-PAGE and electrophoresed onto polyvinylidene difluoride membranes membranes. The membranes were blocked with 5% non-skim milk at room temperature for 2 hours, and incubated with rabbit monoclonal antibodies against KDM1A (1:1,000, Abcam USA, Cambridge, MA, USA) and GAPDH (1:2,000, Abcam USA) at 4°C overnight. HRP-conjugated secondary antibody (1:1,000, OriGene Technologies, Inc., Beijing, People’s Republic of China) was used to incubate the membranes for 30 minutes at room temperature. Finally, signals were detected by the enhanced chemiluminescence kit (BestBio, Shanghai, People’s Republic of China) for 3–5 minutes at room temperature.
Flow cytometric analysis
At 48 hours following transfection, 2×106 single cells were harvested with PBS and fixed in 70% ethanol overnight. Next day, cells were washed with PBS and incubated in propidium iodide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and Annexin V-FITC (BestBio) at 37°C for 15 minutes. Cell apoptosis was evaluated using a BD FACSCanto II analyzer with Mac G4 Computer Workstation and Cell Quest Softwares (BD Biosciences, San Jose, CA, USA). Three independent experiments were repeated.
MTT assay for cell proliferation
At 24 hours following transfection, 2×103 single cells were cultured in 96-well plates at 37°C in 5% CO2. Plates were incubated with MTT reagents (5 mg/mL, 20 µL) for 4 hours. Then, dimethyl sulfoxide (150 µL) was used to stop the reaction and the OD was measured at 490 using a microplate reader. The experiment was followed for 5 days and repeated three times.
Statistical analysis
All data are presented as the mean ± SD, and analyzed using SPSS 19.0 software (IBM Corporation, Armonk, NY, USA). The difference between the two groups was analyzed by independent samples t test. All tests were two-sided and P<0.05 was considered to indicate a statistically significant difference.
Results
miR-101 expression levels in DLBCL and correlation with clinicopathological features
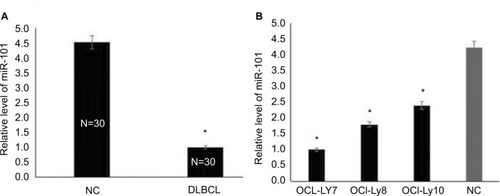
To evaluate the role of miR-101 in DLBCL, the expression levels of miR-101 were examined in DLBCL and normal lymph node tissues using qRT-PCR. As shown in , the expression levels of miR-101 were significantly reduced in DLBCL tissues compared with those in normal controls (P<0.001). Then, the expression levels of miR-101 were also detected in DLBCL cell lines. Results show that the expression levels of miR-101 in DLBCL cell lines were also significantly reduced compared with the average levels of miR-101 in normal lymph node tissues (). These data indicated that miR-101 down-regulation was associated with the formation of DLBCL.
Figure 1 miR-101 had reduced expression levels in DLBCL.
Notes: (A) miR-101 expression levels were measured in DLBCL and normal lymph node tissues by qRT-PCR. *Compared with NC, P<0.001. (B) miR-101 expression levels were measured in DLBCL cell lines by qRT-PCR. *Compared with NC, P<0.001.
Abbreviations: DLBCL, diffuse large B cell lymphoma; NC, normal control.
To further investigate the clinical significance of miR-101 in DLBCL, the correlation between miR-101 expression and clinicopathological features was analyzed. The median value of miR-101 expression in DLBCL tissues was regarded as the cutoff, and then miR-101 expression was divided into high-expression and low-expression. As shown in , miR-101 low expression was positively associated with tumor size (P=0.034), clinical stage (P=0.013) and IPI scores (P=0.026). Thus, these data indicated that miR-101 down-regulation might contribute to the progression of DLBCL.
Table 1 The correlation between miR-101 expression and clinicopathological features was analyzed in DLBCL
miR-101 down-regulation regulates cell apoptosis and proliferation in DLBCL
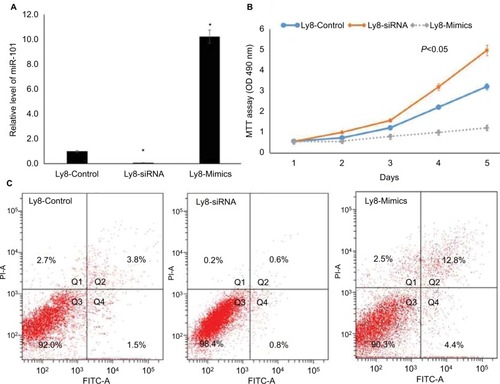
Next, the OCL-LY8 cell line was selected to knock-down and up-regulate the expression of miR-101, respectively. As shown in , miR-101 expression was successfully knocked-down by transferring miR-101 siRNA (P<0.001). After transfection with miR-101 mimics, miR-101 expression was significantly up-regulated (P<0.001) compared with controls. MTT assays revealed that miR-101 down-regulation significantly promoted cell proliferation, while miR-101 up-regulation received a reverse effect (). Flow cytometric analysis revealed that miR-101 down-regulation significantly inhibited cell apoptosis, and miR-101 up-regulation received a reverse effect ().
Figure 2 miR-101 down-regulation regulated cell apoptosis and proliferation in DLBCL.
Notes: (A) The down-regulation and up-regulation of miR-101 in OCL-LY8 cells were validated by qRT-PCR. *Compared with control, P<0.001. (B) MTT assay revealed that miR-101 regulated cell proliferation. (C) Flow cytometric analysis revealed that miR-101 regulated cell apoptosis.
KDM1A is the target of miR-101 to regulate cell proliferation and apoptosis
To investigate the mechanism of miR-101 in DLBCL, Tar-getScanHuman 7.1 was used to predict the targets of miR-101. The predicted consequential pairing of the target region of miR-101 was located at position 166–173 of KDM1A (), and KDM1A was predicted as the possible target of miR-101. To validate the prediction, KDM1A expression was detected in DLBCL and normal lymph node tissues by qRT-PCR. As shown in , KDM1A expression levels in DLBCL tissues were significantly higher compared with those in normal lymph node tissues (P<0.001). KDM1A high-expression was significantly correlated with tumor size, extra-nodal status and IPI (, P<0.05). Moreover, KDM1A expression was adversely correlated with the expression of miR-101 (R=−0.577, P=0.004). When miR-101 expression was successfully down-regulated and up-regulated in OCL-LY8 cells, the mRNA and protein expression levels of KDM1A were detected. Results show that miR-101 down-regulation resulted in a significant increase in KDM1A mRNA level and miR-101 up-regulation reversed the effect (). Meanwhile, miR-101 down-regulation increased the expression of KDM1A protein, which was reversed by miR-101 up-regulation (). The expression of KDM1A was significantly knocked down in OCL-LY8 cells, while the change of miR-101 expression level wasn’t observed compared with control (). These data indicated that KDM1A was the target of miR-101.
Figure 3 KDM1A is the target of miR-101.
Notes: (A) KDM1A was the predicted target of miR-101 by TargetScanHuman 7.1. (B) KDM1A was highly expressed in DLBCL tissues compared with those in normal tissues. *Compared with NC, P<0.001. (C) miR-101 negatively regulated the mRNA level of KDM1A. *Compared with control, P<0.001. (D) miR-101 negatively regulated the protein level of KDM1A.
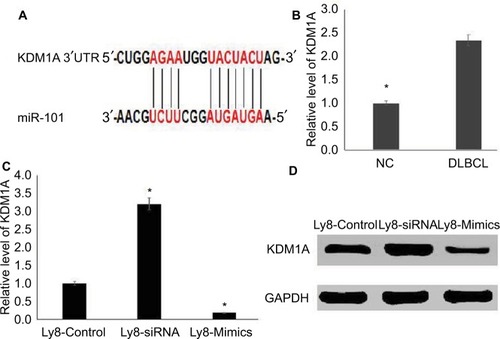
Figure 4 miR-101 negatively regulated KDM1A to affect cell proliferation and apoptosis.
Notes: (A) KDM1A down-regulation didn’t affect the expression of miR-101. *Compared with control, P<0.001. (B) MTT assay shows that KDM1A down-regulation resulted in the inhibition of cell proliferation and partially enhanced the miR-101-induced inhibition of cell proliferation. (C) Flow cytometric analysis revealed that KDM1A down-regulation promoted cell apoptosis and partially enhanced the miR-101-induced promotion of cell apoptosis.
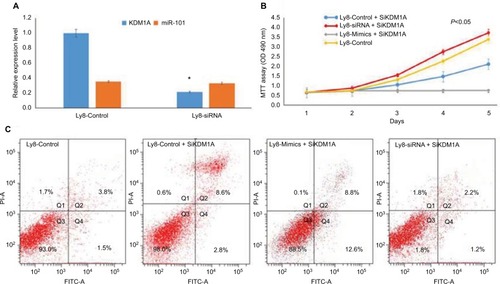
To investigate the role of KDM1A in cell proliferation and apoptosis, KDM1A siRNA and miR101 siRNA/mimics were co-transferred into OCL-LY8 cells, respectively. MTT assay showed that KDM1A down-regulation resulted in the inhibition of cell proliferation (). Moreover, KDM1A knock-down in miR-101-expressing cells partially enhanced the miR-101-induced inhibition of cell proliferation () and promotion of cell apoptosis (). These data suggested that KDM1A was a functionally relevant target of the effect of miR-101 on cell proliferation and apoptosis.
Table 2 The correlation between KDM1A expression and clinicopathological features was analyzed in DLBCL
Discussion
DLBCL is an aggressive disease and is characterized by heterogeneous morphology, clinical presentations, molecular and genetic features.Citation17,Citation18 The pathogenesis of DLBCL is complicated, being involved in different genetic and epigenetic changes.Citation19,Citation20 Recently, miRNAs were reported to be associated with the formation and development of DLBCL.Citation6,Citation7 miR-101 is reported to be associated with tumor formation and plays key roles in cell apoptosis, proliferation, migration and invasion.Citation11–Citation17 However, the role of miR-101 in DLBCL is unknown. Meanwhile, the effect of miR-101 on cell proliferation and apoptosis in DLBCL is unknown.
In this study, it was observed that miR-101 was down-regulated in DLBCL tissues compared with normal tissues. The same trend was recorded in DLBCL cell lines. In addition, miR-101 downregulation was correlated with clinical stage, tumor size and IPI scores. These data indicate that miR-101 downregulation might contribute to the formation and progression of DLBCL. Meanwhile, Chen et alCitation21 reported that miR-101 was down-regulated in gallbladder carcinoma patients and correlated with tumor size, tumor invasion, lymph node metastasis, TNM stage and poor survival. Han et alCitation22 miR-101 downregulation positively correlated with lymph node metastasis and poor prognosis in non-small-cell lung cancer. Thus, our data seem to be in line with the current findings that miR-101 down-regulation correlates with tumor formation and progression.
Then, the biological functions of miR-101 in DLBCL were further investigated. Our data revealed that miR-101 played a key role in regulating cell proliferation and apoptosis. miR-101 downregulation promoted cell proliferation and inhibited cell apoptosis, which further supported that miR-101 was a tumor suppressor in DLBCL. Studies had confirmed that the role of miRNAs in tumors relied on their downstream targets.Citation23,Citation24 So, TargetScanHuman was used to predict the candidate target of miR-101 and KDM1A was confirmed to be the specific target of miR-101. miR-101 directly regulated the expression of KDM1A. Currently, KDM1A is reported to be highly expressed in some types of tumor and KDM1A up-regulation was connected with tumor formation and progression.Citation25–Citation27 However, the expression of KDM1A in DLBCL was unclear. Our data showed that KDM1A was highly expressed in DLBCL tissues compared with those in normal tissues, which was adversely correlated with the expression of miR-101. KDM1A knock-down in DLBCL cells significantly inhibited cell proliferation and promoted cell apoptosis. KDM1A siRNA attenuated the promoting effect of miR-101 down-regulation on DLBCL cell proliferation, while enhancing the suppressing effect of miR-101 up-regulation on DLBCL cell apoptosis. These data suggest that miR-101 negatively regulated KDM1A to affect cell proliferation and apoptosis. Moreover, Zhang et alCitation28 reported that miR-101 inhibited cell proliferation and promoted apoptosis in endometrial cancer by regulating PI3K/Akt/mTOR. Lin et alCitation14 reported that miR-101 suppressed cell proliferation and induced cell apoptosis by targeting EZH2 in esophageal cancer. Bao et alCitation29 reported that miR-101 inhibited the proliferation and metastasis in gallbladder carcinoma by regulating the MAPK/Erk and Smad pathways.
In conclusion, this study is the first to report that miR-101 expression is down-regulated in DLBCL and correlates with tumor formation and progression. Moreover, miR-101 regulates cell proliferation and apoptosis by targeting KDM1A in DLBCL cells.
Data sharing statement
Data are available from the corresponding author upon reasonable request.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Acknowledgments
We would like to thank all patients who supported this study. This study was supported by the Fujian Provincial Natural Science Foundation (no. 2018J01203) and Joint Funds for the innovation of science and Technology, Fujian province (Grant number: 2017Y9070).
Disclosure
The authors report no conflicts of interest in this work.
References
- HeSMiaoXWuYUpregulation of nuclear transporter, Kpnβ1, contributes to accelerated cell proliferation- and cell adhesion-mediated drug resistance (CAM-DR) in diffuse large B-cell lymphomaJ Cancer Res Clin Oncol2016142356157226498772
- ZangCEuckerJLiuHInhibition of pan-class I phosphatidyl-inositol-3-kinase by NVP-BKM120 effectively blocks proliferation and induces cell death in diffuse large B-cell lymphomaLeuk Lymphoma201455242543423721513
- ChenFLiuSZhouYShenHZuoXMAD2 overexpression is associated with high cell proliferation and reduced disease-free survival in primary gastrointestinal diffuse large B-cell lymphomaHematology201621739940327077767
- ObermannECWentPZimpferAExpression of minichromosome maintenance protein 2 as a marker for proliferation and prognosis in diffuse large B-cell lymphoma: a tissue microarray and clinicopathological analysisBMC Cancer2005516216368013
- FarinaFMInguscioAKunderfrancoPCortesiAEliaLQuintavalleMMicroRNA-26a/cyclin-dependent kinase 5 axis controls proliferation, apoptosis and in vivo tumor growth of diffuse large B-cell lymphoma cell linesCell Death Dis201786e289028640256
- FanQMengXLiangHmiR-10a inhibits cell proliferation and promotes cell apoptosis by targeting BCL6 in diffuse large B-cell lymphomaProtein Cell201671289991227815824
- DiaoXZhouJWangSMaXUpregulation of miR-132 contributes to the pathophysiology of COPD via targeting SOCS5Exp Mol Pathol2018105328529230292646
- ChenGZhouHmiRNA-708/CUL4B axis contributes into cell proliferation and apoptosis of osteosarcomaEur Rev Med Pharmacol Sci201822175452545930229816
- LuKShenHZhuSBiSWuSEffects of miRNA-130a on the proliferation and apoptosis of glioma cell linesOncol Lett20181622478248230013640
- CaoKLiJZhaoYmiR-101 inhibiting cell proliferation, migration and invasion in hepatocellular carcinoma through downregulating GirdinMol Cells20163929610226743900
- YanKTianJShiWXiaHZhuYlncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3pCell Physiol Biochem2017423999101228683446
- ShengYLiJZouCDownregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5aArch Virol201415992397241024788845
- LuoCMerzPRChenYmiR-101 inhibits melanoma cell invasion and proliferation by targeting MITF and EZH2Cancer Lett2013341224024723962556
- LinCHuangFLiQZZhangYJmiR-101 suppresses tumor proliferation and migration, and induces apoptosis by targeting EZH2 in esophageal cancer cellsInt J Clin Exp Pathol20147106543655025400732
- SmitsMNilssonJMirSEmiR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesisOncotarget20101871072021321380
- BuechnerJTømteEHaugBHTumour-suppressor microR-NAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastomaBr J Cancer2011105229630321654684
- WuXZhouJWuZmiR-101-3p suppresses HOX transcript antisense RNA (HOTAIR)-induced proliferation and invasion through directly targeting SRF in gastric carcinoma cellsOncol Res20172581383139028251884
- SnakYIndrawatiIdayatiKWArfianNAnggorowatiNMolecular subtypes, apoptosis and proliferation status in Indonesian diffuse large B-cell lymphoma casesAsian Pac J Cancer Prev201819118519129373912
- MiaoXWuYWangYY-box-binding protein-1 (YB-1) promotes cell proliferation, adhesion and drug resistance in diffuse large B-cell lymphomaExp Cell Res2016346215716627397581
- SunXLiBXieBDCZ3301, a novel cytotoxic agent, inhibits proliferation in diffuse large B-cell lymphoma via the STAT3 pathwayCell Death Dis2017810e311129022919
- ChenFLiuSZhouYShenHZuoXMAD2 overexpression is associated with high cell proliferation and reduced disease-free survival in primary gastrointestinal diffuse large B-cell lymphomaHematology201621739940327077767
- HanLChenWXiaYmiR-101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E-box binding homeobox 1Am J Transl Res20181041172118329736210
- ChenJSunDChuHScreening of differential microRNA expression in gastric signet ring cell carcinoma and gastric adenocarcinoma and target gene predictionOncol Rep20153362963297125964059
- ProfumoVDoldiVGandelliniPZaffaroniNTargeting microRNAs to withstand cancer metastasisMethods Mol Biol2015121841543725319666
- Bennani-BaitiIMMachadoILlombart-BoschAKovarHLysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing’s sarcoma, osteosarcoma, and rhabdomyosarcomaHum Pathol20124381300130722245111
- KashyapVAhmadSNilssonEMThe lysine specific demethyl-ase-1 (LSD1/KDM1A) regulates VEGF-A expression in prostate cancerMol Oncol20137355555623384557
- CaiLChenQFangSLianMCaiMMicroRNA-329 inhibits cell proliferation and tumor growth while facilitates apoptosis via negative regulation of KDM1A in gastric cancerJ Cell Biochem201811943338335129130516
- ZhangSWangMLiQZhuPmiR-101 reduces cell proliferation and invasion and enhances apoptosis in endometrial cancer via regulating PI3K/AKT/mTORCancer Biomark201721117918629081412
- BaoRFShuYJHuYPmiR-101 targeting ZFX suppresses tumor proliferation and metastasis by regulating the MAPK/ERK and Smad pathways in gallbladder carcinomaOncotarget2016716223392235426968949
